Potential microbial contamination from drilling lubricants into subseafloor rock cores
H. Lizethe Pendleton
Katrina I. Twing
Shahrzad Motamedi
William J. Brazelton
International Ocean Discovery Program (IODP) Expedition 357: “Serpentinization and Life” drilled shallow cores into the Atlantis Massif near the Mid-Atlantic Ridge in October 2015 using seabed drills. Serpentinization and other geochemical processes occurring within the Atlantis Massif release hydrogen, methane, and other chemicals that can potentially fuel microorganisms through chemosynthesis. The subseafloor rock cores collected during IODP Exp. 357 are the first of their kind, meaning the analysis and interpretation of these samples required new methodologies, including a specialized approach for distinguishing endemic subsurface inhabitants from potential contaminants from various sources. Background samples of various potential contamination sources were collected during sampling: 109 samples of seawater collected before, during, and after drilling; 20 samples of greases and oils associated with the drilling equipment; and samples of the laboratory's ambient air. Despite the widespread usage of drilling lubricants and the importance of controlling contamination in drill-core samples for microbiological analyses, no studies to date have looked at DNA in drilling greases and oils. In this study, drilling lubricants were analyzed as possible sources of microbial contamination of subseafloor rock core samples by environmental sequencing of 16S rRNA genes. We find that microbial signatures from drilling lubricants are only found in low abundance in seafloor samples (at most a few percent of total sequence counts), with laboratory contaminants being a greater source of contamination.
- Article
(1209 KB) - Full-text XML
-
Supplement
(14195 KB) - BibTeX
- EndNote
Due to the low biomass of many subsurface habitats (Kallmeyer, 2017; Smith et al., 2000b), there is a need for improved quality control metrics in order to distinguish between endemic microbial communities and those introduced through contamination (Friese et al., 2017; Smith et al., 2000b; Wilkins et al., 2014; Kallmeyer 2017; Yanagawa et al., 2013; Santelli et al., 2010; Smith et al., 2000b). Contamination of drill core samples can occur from multiple sources before, during, and after drilling. Tracers introduced during drilling (e.g., Friese et al., 2017; Kallmeyer, 2017; Kallmeyer et al., 2006; Lever et al., 2006; Smith et al., 2000a; Yanagawa et al., 2013) are an essential tool for tracking environmental contamination that occurs during drilling, but such tracers cannot identify all possible sources of contamination. Methods for tracking and monitoring the level of contamination introduced during drilling can be generally grouped into three categories: (1) particle tracers (e.g., microspheres), (2) chemical or dissolved tracers (e.g., perfluorocarbon compounds such as perfluoromethylcyclohexane – PFC), and (3) microbiological analyses (e.g., 16S rRNA, fatty acids) (Kallmeyer et al., 2006). Depending on the choice of tracer, various techniques can be used to determine the level of contamination of the drill core samples.
The Lost City is an iconic hydrothermal vent system located on the Atlantis Massif, near the Mid-Atlantic Ridge and Atlantis Fracture Zone (Kelley et al., 2005). The Atlantis Massif is a site of active seafloor serpentinization, and hydrothermal fluids venting through the Lost City chimneys contain products of serpentinization reactions, including hydrogen and methane gas, that can fuel chemoautotrophic microorganisms (Kelley et al., 2005; Lang and Brazelton, 2020). During International Ocean Discovery Program (IODP) Expedition 357: “Serpentinization and Life”, shallow rock cores were drilled in several locations along the Atlantis Massif to recover serpentinite cores where serpentinization is actively occurring, using seabed drills (Früh-Green et al., 2018). Seabed drills differ from traditional platform drilling as they are lowered directly to the drilling site and use bottom seawater as the drilling fluid instead of fluid from a non-indigenous source (Freudenthal and Wefer, 2007). Two seabed drills were used on the Royal Research Ship James Cook for this expedition: RD2 (British Geological Survey) and MARUM-MeBo70 (Center for Marine Environmental Sciences at the University of Bremen) (Früh-Green et al., 2017a; Freudenthal and Wefer, 2007). IODP Exp. 357 employed the use of the synthetic tracer PFC (Smith et al., 2000b) mixed into flushing seawater in an effort to assess the level of contamination introduced into the cores (Orcutt et al., 2017).
The subseafloor rock cores collected during IODP Exp. 357 are the first oceanic crust samples to be collected with seabed drills and suitable for microbiology, so the analysis and interpretation of these samples required the development of new methodologies, including a specialized approach for distinguishing true subsurface microbial inhabitants from surface contaminants (Motamedi et al., 2020). In addition to investigating seawater and lab air as potential sources of contamination of the core samples, the industrial oils and greases used during the drilling process and which potentially came into direct contact with the core samples were previously examined as potential sources of organic molecule contaminants (Hickok et al., 2018). Here, we report an investigation of potential microbial contamination in these same greases and oils, as measured by DNA sequencing.
2.1 Sample collection and processing
Drill core and seawater samples were collected during IODP Exp. 357 and are described in detail in Motamedi et al. (2020). Briefly, core subsamples identified for microbiological analyses were immediately retrieved from the drill upon its return to the ship deck, wrapped in acid-washed and autoclaved teflon, and stored at −80 ∘C. Core samples were then shipped to the Kochi Core Center (Japan) for further processing and subsampling under sterile conditions (Früh-Green et al., 2017b; Orcutt et al., 2017). Please see Früh-Green et al. (2017a, 2018) for more details on lithology and other details of the recovered cores.
Prior to the deployment of the drill at each site, the ship's conductivity, temperature, and depth (CTD) Niskin bottle rosette was cast to collect a water-column profile, with six 10 L bottles being triggered approximately 2–3 m above the seafloor and three 10 L bottles collected at even intervals throughout the water column (Früh-Green et al., 2017b). Additionally, a 4 L bucket was used to collect surface seawater surrounding the ship. Ten-liter Niskin bottles were also attached to each drill and were used to collect bottom seawater during and immediately after drilling. A total of 109 seawater samples were collected during IODP Exp. 357. All seawater samples were filtered with a peristaltic pump through 0.22 µm Sterivex™ filters onboard the ship and stored at −80 ∘C until DNA extraction (Motamedi et al., 2020).
The drill grease and oil samples were collected in sterile 15 mL tubes directly from their original product packaging. Additionally, one methanol-soaked filter used to wipe down a stainless-steel core liner, plastic shards shaved from a plastic core liner, and samples of spray paint that were used on the drill were collected. These samples were all stored at −80 ∘C until extraction. Two sets of these samples were collected, one for testing of organic chemical contamination (Hickok et al., 2018) and the other for microbial contamination testing, described here. All grease/oil extractions took place in a HEPA-filtered room using an aseptic technique. The lab bench was wiped down with 80 % ethanol prior to each set of extractions. Lab air samples were collected by filtering air through 0.1 µm Puradisc 25 mm PTFE syringe filters (GE Healthcare Whatman, Pittsburgh, Pennsylavania, USA) by a dual-head Air Cadet Model 420-2901-00FK (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The lab air was vacuumed through a total of three different filters for 9 h each and was combined during DNA extraction. DNA extraction and purification were performed using the same protocols and reagents as outlined in Motamedi et al. (2020).
2.2 DNA extraction and purification
Test DNA extractions were performed to determine the best protocol for use on the industrial grease and oil samples. For the test samples, LMX “Red” Grease (Plews & Edelmann, IL, USA), WD-40 spray (CA, USA), and mineral oil were selected for their accessibility and similarity to industrial grade oils and greases used during IODP Exp. 357. The quality and quantity of extracted DNA from six different protocols were evaluated (Table 1). Two sets of extractions were performed: the first set were unaltered test samples and the second set were spiked with 10 µL of a turbid suspension of E. coli cells to assess DNA recovery. Once the samples were extracted, both sample sets underwent DNA purification via 2x SPRI beads (Rohland et al., 2012). After each step in the purification process, the samples were quantified with a Qubit fluorometer (Invitrogen, CA, detection limit of 50 ng mL−1) and NanoDrop spectrophotometer (Thermo Fisher Scientific, MA) to assess DNA quantity and quality (Table 1). Of the six DNA-extraction methods, the FastDNA® SPIN Kit (Qbiogene Inc., CA) was the only method to yield DNA from all three test samples.
Table 1Comparison of DNA-extraction methods with three test samples. Six DNA-extraction methods were compared on three test samples (WD-40, mineral oil, and LMX “Red” grease), chosen for their similar properties to industrial grade lubricants. The test samples were spiked with E. coli cells. All DNA yields from un-spiked test samples were below the detection limit (BDL). Each column shows the total amount of DNA extracted (ng).
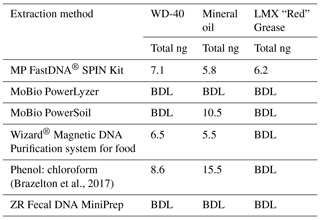
Table 2Description and DNA concentration of drill-associated samples. Twenty samples were taken during the duration of the cruise, including grease, oil, plastic shards, spray paint, and a cotton filter liner. Total DNA (ng) values represent measurements post DNA purification.
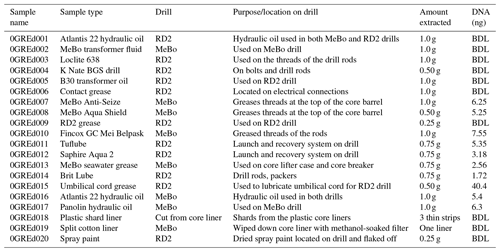
DNA was isolated from the Exp. 357 grease and oil samples using the FastDNA® SPIN Kit according to manufacturer recommendations, except that a Mini-Beadbeater-16 (Biospec Products, OK) was used in place of the FastPrep® instrument. The quantity of starting material for the extraction ranged from 0.25 to 1.0 g (Table 2). Extracted DNA was purified via 2x SPRI beads (Rohland et al., 2012). If replicate extractions of the same sample were possible, the extracted DNA from those replicates was pooled together during the DNA purification step. Additionally, six blank samples of Invitrogen UltraPure™ distilled water were extracted alongside the grease and oil samples as an additional precaution to test for kit contamination, potential contamination introduced during the extraction process, and sequencing contamination (Salter et al., 2014). The extraction of DNA from rock core, seawater, and laboratory air samples was previously described in detail in Motamedi et al. (2020).
2.3 Sequencing and analysis of 16S rRNA genes
Purified DNA preparations from IODP Exp. 357 rock cores, seawater, laboratory air, and greases and oils were sent to the Michigan State University Research and Technology Support Facility Genomics Core for sequencing of the V4 region of the 16S rRNA gene using the duel-indexed Illumina fusion primers 515F-806R (Kozich et al., 2013). All grease and oil samples were submitted for sequencing twice (i.e., sequencing replicates), and the results from both replicates are included in our analysis, with the exception of a single sample, GREd003, one replicate of which was determined to have been compromised during sequencing. Sequences from seawater and rock core samples were previously reported in Motamedi et al. (2020). Analysis of the 16S rRNA gene amplicon sequences from the greases and oils, rock cores, seawater, laboratory air, and extraction blanks was conducted with the mothur (v.1.39.5) software platform (Schloss et al., 2009). Sequences with > 8 homopolymers and > 0 ambiguous bases were removed from downstream analyses, and the sequences were then pre-clustered with the mothur command pre.cluster (diffs = 1) to remove rare sequences most likely created by sequencing errors (Schloss et al., 2011). Operational taxonomic units (OTUs) were formed with a 97 % similarity threshold using the VSEARCH DGC clustering algorithm (Rognes et al., 2016) in mothur. Of the 31 406 783 paired sequences, 75 189 OTUs were identified among the greases and oils, seawater, rock cores, laboratory air, and extraction blanks. Taxonomic classification of all OTUs was performed with mothur using the SILVA reference alignment (SSURefv132) and taxonomy outline (Pruesse et al., 2012). The proportion of contamination from seawater, laboratory air, or industrial grease and oil into each rock core sample was estimated using SourceTracker2 v.2.0.1 (Knights et al., 2011). All sequence data are available via NCBI SRA BioProject PRJNA575221.
3.1 DNA-extraction tests for industrial greases and oils
To our knowledge, the extraction of DNA from industrial greases and oils has not been previously published, either in the context of scientific drilling projects or in other uses of industrial lubricants. DNA-extraction protocols have been published for testing the integrity of food oils such as olive oil (Busconi, et al., 2003; Consolandi, et al., 2008; Testolin and Lain, 2005) and soybean oil (Pauli et al., 1998), but not industrial oils. Multiple DNA-extraction protocols were evaluated for this study using three test samples (LMX “Red” Grease, WD-40 spray, and mineral oil) that were spiked with E. coli cells before DNA extraction. The MPBio FastDNA® SPIN Kit (Qbiogene, Inc., CA) was the only method able to extract detectable DNA from all three of the test samples (Table 1). Extractions from these test samples highlighted viscosity as a key challenge for adapting extraction protocols for greases and oils. In general, less viscous oils were easier to extract than the more viscous grease samples due to difficulties in implementing the physical lysing of thick greases. 16S rRNA gene amplicon sequencing of the spiked test samples confirmed that the recovered DNA was dominated by E. coli (data not shown).
3.2 Microbial composition of grease and oil samples
To assess the potential of drilling equipment to introduce contamination into drill core samples, we collected 20 samples of greases, oils, plastic shards, spray paint, and a cotton filter that wiped a stainless-steel core liner, all of which were used during IODP Exp. 357 (Table 2). All materials that were sampled had some interaction with the drills and introduced a potential for contamination into the rock cores. Six samples of Invitrogen UltraPure™ distilled water were extracted alongside the grease and oil samples to account for any possible contamination introduced from the DNA-extraction kit or during the extraction and sequencing process.
From these 26 samples, we obtained a total of 4 339 588 paired sequences of 16S rRNA gene amplicons, which were clustered into 5629 OTUs at a 97 % sequence similarity threshold. Any OTUs detected in the DNA-extraction blank samples were removed from the dataset, leaving 4694 OTUs (Table S1 in the Supplement). Gammaproteobacteria constituted the highest percentage of taxa in the oil and grease samples (32 % of total sequences; Fig. 1), and the most abundant Gammaproteobacteria OTUs could not be classified below the class level (Table S1). Alphaproteobacteria (17 % of total sequences) were primarily represented by Sphingomonadaceae, Rhodobacteraceae, and Acetobacteraceae. Bacteroidia (10 % of total sequences) were primarily represented by Flavobacteriaceae, Spirosomaceae, and Hymenobacteraceae. Betaproteobacteriales (4 % of total sequences, but note that Betaproteobacteriales are classified as an order within class Gamamproteobacteria in the SILVA taxonomy) were dominated by various genera of the Burkholderiaceae.
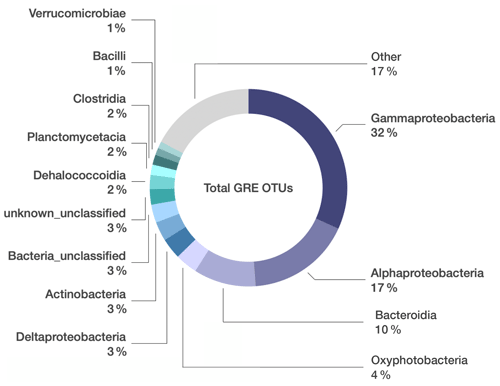
Figure 1Microbial composition unique to grease and oil samples used on seafloor drills. Relative abundance of bacterial classes identified in grease and oil samples. Sequences that were also present in the controls (extraction kit blank, lab air, and MilliQ water) were manually removed from the dataset. “Other” includes 95 additional classes whose relative abundance was < 1 % of the total community composition.
DNA sequences in the oils and greases had high similarity to sequences from a wide range of environments, including soil (e.g., NCBI accessions HM104622, MG716681, AM940870, KP786168, NR_163645), glaciers (HQ333317, MN880348), lake sediments (MT067094), a geothermal plant (KY077452), a shallow marine hydrothermal vent (GU369930), and seawater (JN233022, KX177824). Sequences associated with petroleum-contaminated environments were also identified (EU328045, KY190357). Many of the matching sequences were associated with drilling projects, such as an IODP borehole (KR072759), marine sediments (CP004387, MF977474), a continental borehole (KP901594), and groundwater wells (KC606558). A few OTUs obtained from a swipe of a core liner barrel matched those from continental subsurface studies (MT067098, HM185963, HM641526). Notably, several OTUs that were abundant in the grease that is used on RD2's umbilical cord were nearly identical to clones from a deep-sea drilling and coring contamination study (Yanagawa et al., 2013), notably including those recovered from the drilling fluid in that study (e.g., AB824901). A summary of best sequence matches is provided in the Supplement (Table S2).
3.3 Potential oil and grease contamination of seawater samples
All of the most abundant grease/oil OTUs were also identified in samples of seawater collected during the expedition. Because the greases and oils were sampled directly from their commercial product packages (except for the core liner swab and paint chips), contamination from seawater into the grease/oil samples seems unlikely. The grease/oil OTUs that are most abundant in seawater samples were almost exclusively derived from GREd008 (AquaShield, lubrication for MeBo core barrel threads) and GREd015 (RD2 umbilical cord grease). Furthermore, seawater samples collected with Niskin bottles mounted on MeBo and RD2 were especially likely to contain OTUs from the AquaShield grease (Table S1). However, overall, these potential grease/oil contaminants represent a small fraction of the total sequence dataset from seawater (< 1 % of all sequence counts).
3.4 Minimal oil and grease contamination of rock cores
The rock core samples collected during IODP Exp. 357 were exposed to potential contamination sources before, during, and after drilling (Fig. 2). The extent of DNA contamination from seawater into the rock cores was investigated by Motamedi et al. (2020), and here, we extend that analysis to include grease and oil samples as additional potential sources of contamination.
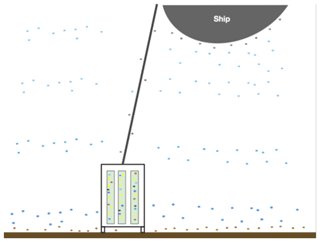
Figure 2Illustration of various microbial communities present in the sampling environment. The different colored dots represent microbes from varying depth ranges in the water column (shades of blue) as well as microbes surrounding the ship, drill, and drill line (grey), microbes at the water–sediment interface, and the actual subsurface microbes (yellow) that live in the rock cores. The blue, grey, and brown dots represent the potential for contamination to the subsurface rock cores and illustrate the need for rigorous contamination testing.
Of the 4694 OTUs identified in all grease and oil samples, 565 OTUs were also identified in at least one rock core sample from IODP Exp. 357 (Table S3). However, 86 of these OTUs were also identified in samples of the ambient lab air, suggesting that some of these sequences represent general contamination from dust particles during laboratory handling. In addition, the taxonomic classifications of many of these OTUs suggest that they are derived from commercial reagents (e.g., Burkholderia) or the human microbiota (e.g., Enterobacteriaceae), based on previous studies (Sheik et al., 2018; Salter et al., 2014), even though they were not detected in the extraction blanks or ambient lab air during our study. OTUs that are suspected to be contaminants on the basis of their taxonomic classification are highlighted in (but not removed from) the tables in the Supplement.
The remaining 479 OTUs that represent potential grease/oil contaminants of the rocks (Table S3) comprise 16 % of the total OTUs and 24 % of the total sequence counts in the rock core samples. However, most (90 %) of these sequence counts in rocks are contributed by OTUs that were found in low abundance in our samples of greases and oils (< 100 total counts across all GRE samples), casting doubt that the greases and oils were the source of most of these contaminants into the rock cores. Abundant OTUs from greases and oils were generally very rare in the rock cores.
Table 3Condensed phylogenetic table of S4 showing the 27 most likely contaminant OTUs from the grease and oil samples.
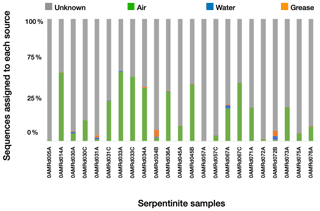
Figure 3Estimated sources of DNA sequences into rock core samples. SourceTracker2 (Knights et al., 2011) was used to identify the most likely source of OTUs into the rock cores. Possible sources were laboratory air (green), seawater (blue), or drilling grease or oil (orange). OTUs with an “unknown” source could not be assigned to a single source and may represent rock-hosted microbes.
We estimated the proportion of DNA sequences from each rock core sample that could be attributed to each potential source of contamination (i.e., seawater, laboratory air, or drill grease and oil) using SourceTracker2 (Fig. 3). OTUs with “unknown” sources could not be assigned to a single contamination source and may represent true inhabitants of the rock cores. Lab air was the largest source of contamination into the rock cores, and contamination from other sources was minimal. Greases and oils were estimated to contribute at most a few percent of the sequences in each rock core sample, and their contribution was nearly zero in many of the samples. Nevertheless, the detectable levels of contamination from grease/oil and seawater are notable, considering the extensive precautions employed during handling and processing of the rock core samples (Früh-Green et al., 2017a, 2018; Hickok et al., 2018; Motamedi et al., 2020). These precautions (including the use of bottom seawater as the drilling fluid, immediate freezing of core samples, and shaving of core exteriors with a sterile rock saw in a dedicated facility) are not practical for many drilling projects, suggesting that these contamination levels may be higher in other studies.
We assembled a final list of 27 likely contaminant OTUs from greases and oils (Table S4 and summarized in Table 3) based on their absence in extraction blanks and lab air and their much higher abundance in greases and oils compared to seawater (i.e., > 5 × greater abundance and > 500 total counts in greases and oils; see Table S3 for numbers). These likely contaminants were mostly derived from GREd001 (hydraulic oil used on both RD2 and MeBo) and GREd015 (RD2 umbilical cord grease) and were also moderately abundant in GREd004 (K Nate grease), GREd006 (electrical contact grease), and GREd008 (AquaShield grease). They represent 8 phyla and 10 classes, with Clostridia the most frequently appearing. As noted in Sect. 3.2, the most abundant of these sequences had high similarity to database sequences reported from a wide range of environments.
The possibility of contaminant DNA introduced by greases and oils associated with drilling equipment had not been previously explored. We have demonstrated that DNA can be detected in industrial greases and oils and that these same DNA sequences can also be found at low levels in low-biomass rock cores and in seawater samples. Nevertheless, our results indicate that, for our study, contamination from greases and oils was much less prevalent compared to contamination during laboratory handling, as measured by DNA extracted from dust particles in ambient lab air. Even though we do not expect greases and oils to be the most important source of contamination in most studies, levels of contamination from different sources will vary according to the particular circumstances of each project. Therefore, we recommend that future studies should monitor potential contamination from greases and oils associated with drilling and sampling equipment.
All sequence data are available via NCBI SRA BioProject PRJNA575221. The data are accessible with the provided accession ID at the following link for the SRA database: https://www.ncbi.nlm.nih.gov/sra/ (last access: 18 March 2021).
The supplement related to this article is available online at: https://doi.org/10.5194/sd-29-49-2021-supplement.
HLP carried out the experiments, analyzed the data, and wrote the manuscript. KIT collected samples, designed and supervised the project, and edited the manuscript. SM provided and analyzed data and edited the manuscript. WJB designed and supervised the project and edited the manuscript.
The authors declare that they have no conflict of interest.
This research depended on data, samples, and technical expertise provided by International Ocean Discovery Program (IODP) Expedition 357 supported by the European Consortium for Ocean Research Drilling (ECORD) and implemented by the ECORD Science Operator (ESO). The shipboard sample-processing protocols were executed by ESO Expedition Project Manager Carol Cotterill, ESO Operations Superintendent David Smith, chief scientists Beth Orcutt and Gretchen Früh-Green, the crews of the R.R.S. James Cook and the MeBo and RD2 seabed drills, and shipboard scientists Susan Lang, Marvin Lilley, Yuki Morono, Marianne Quéméneur, and Matthew Schrenk. We are sincerely grateful to the crews of RD2, led by Michael Wilson, and MeBo, led by Tim Freudenthal, for identifying and providing us with the grease and oil samples. We also thank Susan Lang, Katherine Hickok, and Tran Nguyen for discussion and guidance regarding their previous analyses of the samples and Michael Wilson and Beth Orcutt for helpful comments on the manuscript.
We are grateful to the NASA Rock-Powered Life team and the Deep Carbon Observatory for their support and productive conversations and for an American Society for Microbiology Undergraduate Research Fellowship to H. Lizethe Pendleton.
This research has been supported by the NASA Astrobiology Institute (grant no. CAN-7) and the National Science Foundation-funded US Science Support Program (grant no. OCE 14-50528).
This paper was edited by Tomoaki Morishita and reviewed by two anonymous referees.
Brazelton, W. J., Thornton, C. N., Hyer, A., Twing, K. I., Longino, A. A., Lang, S. Q., Lilley, M. D., Früh-Green, G. L., and Schrenk, M. O.: Metagenomic identification of active methanogens and methanotrophs in serpentinite springs of the Voltri Massif, Italy, PeerJ, 5, e2945, https://doi.org/10.7717/peerj.2945, 2017.
Busconi, M., Foroni, C., Corradi, M., Bongiorni, C., Cattapan, F., and Fogher, C.: DNA extraction from olive oil and its use in the identification of the production cultivar, Food Chem., 83, 127–134, https://doi.org/10.1016/S0308-8146(03)00218-8, 2003.
Consolandi, C., Palmieri, L., Severgnini, M., Maestri, E., Marmiroli, N., Agrimonti, C., Baldoni, L., Donini, P., De Bellis, G., and Castiglioni, B.: A procedure for olive oil traceability and authenticity: DNA extraction, multiplex PCR and LDR – universal array analysis, Eur. Food Res. Technol., 227, 1429–1438, https://doi.org/10.1007/s00217-008-0863-5, 2008.
Freudenthal, T. and Wefer, G.: Scientific Drilling with the Sea Floor Drill Rig MeBo, Sci. Dril., 5, 63–66, https://doi.org/10.2204/iodp.sd.5.11.2007, 2007.
Friese, A., Kallmeyer, J., Kitte, J. A., Martínez, I. M., Bijaksana, S., and Wagner, D.: A simple and inexpensive technique for assessing contamination during drilling operations, Limnol. Oceanogr.-Meth., 15, 200–211, https://doi.org/10.1002/lom3.10159, 2017.
Früh-Green, G. L., Orcutt, B. N., Green, S. L., Cotterill, C., Morgan, S., Akizawa, N., Bayrakci, G., Behrmann, J.-H., Boschi, C., Brazelton, W. J., Cannat, M., Dunkel, K. G., Escartin, J., Harris, M., Herrero-Bervera, E., Hesse, K., John, B. E., Lang, S. Q., Lilley, M. D., Liu, H.-Q., Mayhew, L. E., McCaig, A. M., Menez, B., Morono, Y., Quéméneur, M., Rouméjon, S., Sandaruwan Ratnayake, A., Schrenk, M. O., Schwarzenbach, E. M., Twing, K. I., Weis, D., Whattam, S. A., Williams, M., and Zhao, R.: Expedition 357 summary, in: Proceedings of the International Ocean Discovery Program Volume 357, edited by: Früh-Green, G. L., Orcutt, B. N., Green, S. L., Cotterill, C., and the Expedition 357 Scientists, International Ocean Discovery Program Publications, College Station, TX, USA, https://doi.org/10.14379/iodp.proc.357.101.2017, 2017a.
Früh-Green, G. L., Orcutt, B. N., Green, S. L., Cotterill, C., Morgan, S., Akizawa, N., Bayrakci, G., Behrmann, J.-H., Boschi, C., Brazelton, W. J., Cannat, M., Dunkel, K. G., Escartin, J., Harris, M., Herrero-Bervera, E., Hesse, K., John, B. E., Lang, S. Q., Lilley, M. D., Liu, H.-Q., Mayhew, L. E., McCaig, A. M., Menez, B., Morono, Y., Quéméneur, M., Rouméjon, S., Sandaruwan Ratnayake, A., Schrenk, M. O., Schwarzenbach, E. M., Twing, K. I., Weis, D., Whattam, S. A., Williams, M., and Zhao, R.: Expedition 357 methods, in: Proceedings of the International Ocean Discovery Program, 357, edited by: Früh-Green, G. L., Orcutt, B. N., Green, S. L., Cotterill, C., and the Expedition 357 Scientists, International Ocean Discovery Program Publications, College Station, TX, USA, https://doi.org/10.14379/iodp.proc.357.102.2017, 2017b.
Früh-Green, G. L., Orcutt, B. N., Rouméjon, S., Lilley, M. D., Morono, Y., Cotterill, C., Green, S., Escartin, J., John, B. E., McCaig, A. M., Cannat, M., Ménez, B., Schwarzenbach, E. M., Williams, M. J., Morgan, S., Lang, S. Q., Schrenk, M. O., Brazelton, W. J., Akizawa, N., Boschi, C., Dunkel, K. G., Quéméneur, M., Whattam, S. A., Mayhew, L., Harris, M., Bayrakci, G., Behrmann, J.-H., Herrero-Bervera, E., Hesse, K., Liu, H.-Q., Ratnayake, A. S., Twing, K., Weis, D., Zhao, R., and Bilenker, L.: Magmatism, serpentinization and life: Insights through drilling the Atlantis Massif (IODP Expedition 357), Lithos, 323, 137–155, https://doi.org/10.1016/j.lithos.2018.09.012, 2018.
Hickok, K. A., Nguyen, T. B., and Lang, S. Q.: Assessment of apolar lipids in subseafloor rocks and potential contaminants from the Atlantis Massif (IODP Expedition 357), Org. Geochem., 122, 68–77, https://doi.org/10.1016/j.orggeochem.2018.05.003, 2018.
Kallmeyer, J.: Contamination Control for Scientific Drilling Operations, Adv. Appl. Microbiol., 98, 61–91, https://doi.org/10.1016/bs.aambs.2016.09.003, 2017.
Kallmeyer, J., Mangelsdorf, K., Cragg, B., and Horsfield, B.: Techniques for Contamination Assessment During Drilling for Terrestrial Subsurface Sediments, Geomicrobiol. J., 23, 227–239, https://doi.org/10.1080/01490450600724258, 2006.
Kelley, D. S., Karson, J. A., Früh-Green, G. L., Yoerger, D. R., Shank, T. M., Butterfield, D. A., Hayes, J. M., Schrenk, M. O., Olson, E. J., Proskurowski, G., Jakuba, M., Bradley, A., Larson, B., Ludwig, K., Glickson, D., Buckman, K., Bradley, A. S., Brazelton, W. J., Roe, K., Elend, M. J., Delacour, A., Bernasconi, S. M., Lilley, M. D., Baross, J. A., Summons, R. E., and Sylva, S. P.: A Serpentinite-Hosted Ecosystem: The Lost City Hydrothermal Field, Science, 307, 1428–1434, https://doi.org/10.1126/science.1102556, 2005.
Knights, D., Kuczynski, J., Charlson, E. S., Zaneveld, J., Mozer, M. C., Collman, R. G., Bushman, F. D., Knight, R., and Kelley, S. T.: Bayesian community-wide culture-independent microbial source tracking, Nat. Methods, 8, 761–763, https://doi.org/10.1038/nmeth.1650, 2011.
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D.: Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform, Appl. Environ. Microbiol., 79, 5112–5120, https://doi.org/10.1128/AEM.01043-13, 2013.
Lang, S. Q. and Brazelton, W. J.: Habitability of the marine serpentinite subsurface: a case study of the Lost City hydrothermal field, Philos. T. R. Soc. A, 378, 20180429, https://doi.org/10.1098/rsta.2018.0429, 2020.
Lever, M. A., Alperin, M., Engelen, B., Inagaki, F., Nakagawa, S., Steinsbu, B. O., and Teske, A.: Trends in Basalt and Sediment Core Contamination During IODP Expedition 301, Geomicrobiol. J., 23, 517–530, https://doi.org/10.1080/01490450600897245, 2006.
Motamedi, S., Orcutt, B. N., Früh-Green, G. L., Twing, K. I., Pendleton, H. L., and Brazelton, W. J.: Microbial Residents of the Atlantis Massif's Shallow Serpentinite Subsurface, Appl. Environ. Microbiol., 86, e00356-20, https://doi.org/10.1128/AEM.00356-20, 2020.
Orcutt, B. N., Bergenthal, M., Freudenthal, T., Smith, D., Lilley, M. D., Schnieders, L., Green, S., and Früh-Green, G. L.: Contamination tracer testing with seabed drills: IODP Expedition 357, Sci. Dril., 23, 39–46, https://doi.org/10.5194/sd-23-39-2017, 2017.
Pauli, U., Liniger, M., and Zimmermann, A.: Detection of DNA in soybean oil, Z. Lebensm. Unters. Forsch., 207, 264–267, https://doi.org/10.1007/s002170050330, 1998.
Pruesse, E., Peplies, J., and Glöckner, F. O.: SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes, Bioinformatics, 28, 1823–1829, https://doi.org/10.1093/bioinformatics/bts252, 2012.
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F.: VSEARCH: a versatile open source tool for metagenomics, PeerJ, 4, e2584, https://doi.org/10.7717/peerj.2584, 2016.
Rohland, N. and Reich, D.: Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture, Genome Res., 22, 939–946, https://doi.org/10.1101/gr.128124.111, 2012.
Salter, S. J., Cox, M. J., Turek, E. M., Calus, S. T., Cookson, W. O., Moffatt, M. F., Turner, P., Parkhill, J., Loman, N. J., and Walker, A. W.: Reagent and laboratory contamination can critically impact sequence-based microbiome analyses, BMC Biol., 12, 87, https://doi.org/10.1186/s12915-014-0087-z, 2014.
Santelli, C. M., Banerjee, N., Bach, W., and Edwards, K. J.: Tapping the Subsurface Ocean Crust Biosphere: Low Biomass and Drilling-Related Contamination Calls for Improved Quality Controls, Geomicrobiol. J., 27, 158–169, https://doi.org/10.1080/01490450903456780, 2010.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., Lesniewski, R. A., Oakley, B. B., Parks, D. H., Robinson, C. J., Sahl, J. W., Stres, B., Thallinger, G. G., Horn, D. J. V., and Weber, C. F.: Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities, Appl. Environ. Microbiol., 75, 7537–7541, https://doi.org/10.1128/AEM.01541-09, 2009.
Schloss, P. D., Gevers, D., and Westcott, S. L.: Reducing the Effects of PCR Amplification and Sequencing Artifacts on 16S rRNA-Based Studies, PLOS ONE, 6, e27310, https://doi.org/10.1371/journal.pone.0027310, 2011.
Sheik, C. S., Reese, B. K., Twing, K. I., Sylvan, J. B., Grim, S. L., Schrenk, M. O., Sogin, M. L., and Colwell, F. S.: Identification and Removal of Contaminant Sequences From Ribosomal Gene Databases: Lessons From the Census of Deep Life, Front. Microbiol., 9, 840, https://doi.org/10.3389/fmicb.2018.00840, 2018.
Smith, D. C., Spivack, A. J., Fisk, M. R., Haveman, S. A., Staudigel, H., and the Leg 185 Shipboard Scientific Party: Methods for quantifying potential microbial contamination during deep ocean coring, ODP Tech. Note, Ocean Drilling Program, 28, https://doi.org/10.2973/odp.tn.28.2000, 2000a.
Smith, D. C., Staudigel, H., Spivack, A. J., Fisk, M. R., and Haveman, S. A.: Tracer-Based Estimates of Drilling-Induced Microbial Contamination of Deep Sea Crust, Geomicrobiol. J., 17, 207–219, https://doi.org/10.1080/01490450050121170, 2000b.
Testolin, R. and Lain, O.: DNA Extraction from Olive Oil and PCR Amplification of Microsatellite Markers, J. Food Sci., 70, 108–112, https://doi.org/10.1111/j.1365-2621.2005.tb09011.x, 2005.
Wilkins, M. J., Daly, R., Mouser, P. J., Trexler, R., Wrighton, K. C., Sharma, S., Cole, D. R., Biddle, J. F., Denis, E., Fredrickson, J. K., Kieft, T. L., Onstott, T. C., Petersen, L., Pfiffner, S. M., Phelps, T. J., and Schrenk, M. O.: Trends and Future Challenges in Sampling the Deep Terrestrial Biosphere, Front. Microbiol., 5, 481, https://doi.org/10.3389/fmicb.2014.00481, 2014.
Yanagawa, K., Nunoura, T., McAllister, S., Hirai, M., Breuker, A., Brandt, L., House, C., Moyer, C. L., Birrien, J.-L., Aoike, K., Sunamura, M., Urabe, T., Mottl, M., and Takai, K.: The first microbiological contamination assessment by deep-sea drilling and coring by the D/V Chikyu at the Iheya North hydrothermal field in the Mid-Okinawa Trough (IODP Expedition 331), Front. Microbiol., 4, 327, https://doi.org/10.3389/fmicb.2013.00327, 2013.





