Microbial diversity of drilling fluids from 3000 m deep Koyna pilot borehole provides insights into the deep biosphere of continental earth crust
Himadri Bose
Avishek Dutta
Ajoy Roy
Abhishek Gupta
Sourav Mukhopadhyay
Balaram Mohapatra
Jayeeta Sarkar
Sukanta Roy
Sufia K. Kazy
Pinaki Sar
Scientific deep drilling of the Koyna pilot borehole into the continental crust up to a depth of 3000 m below the surface at the Deccan Traps, India, provided a unique opportunity to explore microbial life within the deep granitic bedrock of the Archaean Eon. Microbial communities of the returned drilling fluid (fluid returned to the mud tank from the underground during the drilling operation; designated here as DF) sampled during the drilling operation of the Koyna pilot borehole at a depth range of 1681–2908 metres below the surface (m b.s.) were explored to gain a glimpse of the deep biosphere underneath the continental crust. Change of pH to alkalinity, reduced abundance of Si and Al, but enrichment of Fe, Ca and in the samples from deeper horizons suggested a gradual infusion of elements or ions from the crystalline bedrock, leading to an observed geochemical shift in the DF. Microbial communities of the DFs from deeper horizons showed progressively increased abundance of Firmicutes, Gammaproteobacteria and Actinobacteria as bacterial taxa and members of Euryarchaeota as the major archaeal taxa. Microbial families, well known to strive in strictly anaerobic and extremophilic environments, (e.g. Thermoanaerobacteraceae, Clostridiaceae, Bacillaceae, Carnobacteriaceae, Ruminococcaceae), increased in the samples obtained at a depth range of 2000 to 2908 m b.s. Phylogenetic analysis of common and unique operational taxonomic units (OTUs) of DF samples indicated signatures of extremophilic and deep subsurface relevant bacterial genera (Mongoliitalea, Hydrogenophaga, Marinilactibacillus, Anoxybacillus, Symbiobacterium, Geosporobacter, Thermoanaerobacter). Thermophilic, obligatory anaerobic sulfate-reducing bacterial taxa known to inhabit the deep subsurface were enriched from DF samples using sulfate as a terminal electron acceptor. This report on the geomicrobiology of the DF obtained during drilling of the deep subsurface of the Deccan Traps showed new opportunities to investigate deep life from terrestrial, granite-rock-hosted habitats.
- Article
(11874 KB) - Full-text XML
-
Supplement
(165 KB) - BibTeX
- EndNote
The earth's deep continental crust was found to be of geologically varied morphology with extreme conditions (temperature, pressure, pH etc.) which made it almost impossible for life to survive (Fredrickson and Balkwill, 2006; Colwell and D'Hondt, 2013). Nevertheless, the “deep subsurface” of the earth has been known to host diverse arrays of ecosystems which harbour numerous populations of extremophilic microbial life, comprising a significant (about 19 %) but mostly unexplored parts of earth's biosphere (Whitman et al., 1998; McMahon and Parnell, 2014, 2018; Kieft, 2016; Magnabosco et al., 2018; Soares et al., 2019). In fact, the deep subsurface is among the five main “big habitats” of bacteria and archaea containing about 3×1029 cells (Flemming and Wuertz, 2019). Exploration of deep life within the continental crust mostly aims to provide answers to some of the fundamental questions related to the limits of life on our planet and its evolution, energy mechanisms and metabolism as well as its involvement in cycling of the essential elements of life (Hoehler and Jørgensen, 2013; Breuker et al., 2011; Purkamo et al., 2018; Borgonie et al., 2019).
Deep drilling has been an important method for obtaining subsurface samples for geomicrobiological investigations (Escudero et al., 2018). Generally, for acquiring the sample of more than 300 m depth (especially from crystalline – granite and basalt – rock) a rotary drilling technique has been used which requires the drilling fluid to facilitate the drilling process (Keift et al., 2007; Onstott et al., 1998). Drilling fluid prepared by mixing multiple pre-packaged powdered components, including bentonite and a variety of organic constituents with water to lubricate and cool the drill bit, maintain the hydrostatic pressure during the drilling operations so that intact cores can be retrieved and ultimately carry the rock cuttings back to the surface (Zhang et al., 2006; Keift, 2010). Bentonite (sodium bentonite: aluminium phyllosilicate clay consisting mostly of montmorillonite) has been widely used as drilling fluid in several continental and oceanic drilling operations (Masui et al., 2008; Zhang et al., 2006; Struchtemeyer et al., 2011). Specific advantages of bentonite-based fluids included protection of the drilling tools from corrosion, reduced fluid and gas penetration (Gandhi and Sarkar, 2003), and owing to its characteristic “thixotrophic reaction” it hinders the rock material from dropping back into the drilled shaft (İşçi and Turutoğlu, 2011).
These fluids are essentially considered as potential contaminants to rock cores not only from chemical but also from a biological perspective. Owing to the presence of a variety of complex organic compounds (e.g. xanthan gum, guar gum, glycol, carboxymethylcellulose, polyanionic cellulose (PAC) or starch), diverse heterotrophic microbial populations (bacteria, archaea, fungi) could grow in drilling fluid (Keift et al., 2007; Beeman and Suflita, 1989; Rabia, 1985). A circulatory system in which the fluid remained in continuous use throughout the drilling operation and the scope of the invasion of microorganisms from the upper stratum to the strata below may also pose a matter of further concern (Pedersen et al., 1997; Yanagawa et al., 2013; Smith et al., 2000; Miteva et al., 2014). In spite of the shortcomings, the fluid that returned back to the mud tank has been a subject of geomicrobiological investigation since it can act as a window enabling sampling of the deep subsurface environment and may give us an opportunity to analyse the microbial communities from the geological fluids from faults and fractures (Zhang et al., 2006).
Drilling fluids' microbial diversity as a marker for deep life was studied during a number of drilling operations. Drilling fluid obtained from deep drilling at the Cretaceous-age Piceance basin in Colorado, Triassic-age Taylorsville basin in Virginia, Chinese continental scientific drilling operations (CCSD) at a 240 Ma old ultra-high pressure metamorphic (UHPM) belt located in Jiangsu Province in China and during the first riser drilling operation on the deep-earth research vessel Chikyu was used for microbiological studies (Liu et al., 1997; Roh et al., 2002; Zhang et al., 2005, 2006; Masui et al., 2008). The presence of microorganisms able to survive the extreme oligotrophic conditions of the deep subsurface was detected in these studies. Several strains of an extremely thermophilic, anaerobic chemoorganotroph bacterium Thermoanaerobacter ethanolicus have been previously isolated from the fluids obtained during drilling in the Piceance basin in Colorado and Taylorsville basin in Virginia at different depths of the geological formation (Liu et al., 1997; Roh et al., 2002). This particular organism was also retrieved from the DF samples of CCSD. Extremophilic deep subsurface microorganisms (Caldicellulosiruptor lactoaceticus, Anaerobranca gottschalkii and Sulfolobus solfataricus) were reported from the fluids obtained from the CCSD project (Zhang et al., 2005, 2006). Deep subsurface archaeal populations (Marine Crenarchaeota Group I and II, South African Gold Mine Euryarchaeotic Group (SAGMEG), Soil Group Archaea) were detected in the fluid samples at the time of the first riser drilling operation on Chikyu (Masui et al., 2008). Overall, the microbial ecology of circulating fluids collected from various deep drilling operations (continental and oceanic) showed strong signatures of subsurface microbial populations. Investigating these communities remained scientifically intriguing. Additionally, the drill cuttings along with the fluids also leave behind tons of residue without adequate treatment, generating a large environmental liability. Knowledge about the microbial community of such drilling residue may be useful for developing techniques for drilling mud bioremediation (Guerra et al., 2018).
The present research was undertaken to study the microbial communities present in the returned drilling fluids (designated here as DF) as a proxy for deep granitic subsurface microbial diversity in the Koyna Intra-plate Seismic zone (Deccan Traps, India; https://www.icdp-online.org/projects/world/asia/koyna-india/, last access: 27 January 2020). This unique opportunity to study microbial life within deep terrestrial igneous rocks came to us through scientific deep drilling in this region by the Inter Continental Drilling Programme (ICDP) and Ministry of Earth Sciences (MoES), Government of India, to study the earthquakes. The Deccan Traps, representative of a continental flood basalt province, covers an area of over 0.5 million km2 of lava flows (∼65 Ma) with a total thickness of over 2000 m near the eruptive centre in western India. This thick pile of Deccan basalts has been resting on a ∼2.5 Ga Archaean crystalline basement. With respect to varying temperature, pressure regimes (15 ∘C increase in temperature and 26.7 MPa increase in lithostatic pressure per 1000 m b.s. in granite) and geochemical conditions, this study provided an excellent opportunity to study life in deep igneous terrestrial subsurface (Roy and Rao, 2000). The specific goal of this work was to systematically investigate the microbial diversity of DFs in the depth range of 1681–2908 m b.s. by a culture-independent approach and explore the presence of deep life relevant organisms within these systems. Our results indicated that there was a change in the microbial community structure of DF samples on increasing borehole depth. The phylogenetic positions of unique and shared operational taxonomic units (OTUs) of the DF samples suggest the presence of deep subsurface and extremophilic microbial populations in drilling fluid samples.
2.1 Sampling site and drilling
A 3 km deep pilot borehole (KFD-1) was drilled in the Koyna region (17∘24′6′′ N, 73∘45′8′′ E) of the Deccan Traps, Maharashtra during March–June 2017 under the pilot phase of the Koyna scientific drilling project to comprehend the genesis of earthquakes triggered by artificial water reservoirs in the Koyna intraplate seismic zone. Several modifications to the standard drilling procedure were made in order to collect the core and drilling fluid samples in such a way that they could be later used reliably for geochemical and geomicrobiological analysis. All instruments in contact with the core samples were cleaned properly before use. The drilling fluid (bentonite) was kept in a separate tank and sodium fluorescein (500 mg m−3) was added to the tank before the coring process to evaluate the penetration of drill mud into the core. Drilling fluid samples that returned after circulating to the depths ranging from 1681 to 2908 m b.s. were collected following aseptic techniques along with the rock cores (core 1–core 7) at the time coring at the depths ranging from 1681 to 2908 m b.s. These returned DF samples were designated as DF1 to DF7. Along with that the in-flow fluid (before its introduction in the borehole) was also collected. A list of samples, depth and other physiochemical parameters has been presented in Table S1 in the Supplement. Samples were stored in sterile containers at 4 ∘C for shipment. In laboratory, the samples were stored at −80 ∘C.
2.2 Geochemical analysis
The DF samples (DF1–DF7) were dried overnight in a hot air oven (80 ∘C) to remove the water content. The dried cakes were ground to powder using a mortar and pestle. The powdered DFs were again kept overnight at 80 ∘C to remove any leftover moisture and stored in a vacuum desiccator. The powdered samples were analysed for their elemental composition by energy-dispersive X-ray spectroscopy (EDX) using a field emission scanning electron microscope (Zeiss Merlin) attached to an EDX detector. For pH and conductivity measurements, DF samples were incubated in distilled water overnight at a 1 : 10 ratio (w∕v). All measurements were done using highly sensitive probes fitted with a multiparameter (Orion) (Thermo electron corporation, Beverly, MA) (Islam et al., 2014). Temperature for each depth (at which rock cores and DF samples were collected) was measured on-site during drilling.
2.3 DNA extraction
Microbial diversity and community composition of DFs were studied using an approach based on next-generation sequencing (NGS) and using the extracted DNA from the respective samples. Environmental DNA from DFs (250 mg of DF was taken for each extraction) was extracted in triplicates using the DNeasy® PowerSoil® kit (Qiagen 12888-50) according to the manufacturer's protocol. A blank DNA extraction was performed (no sample) at the same time as a negative control and was used subsequently in the PCR and sequencing steps to check chances of any possible contamination. The quality of the extracted environmental DNA and its concentration was measured using a NanoDrop 2000 spectrophotometer, followed by fluorometric quantitation using Qubit (Thermo-Fisher Scientific).
2.4 Quantitative real time PCR (qPCR)
Quantification of the bacterial and archaeal populations in DFs was performed by estimating the copy number of bacteria- and archaea-specific 16S rRNA genes. Details of the qPCR primers used are provided in Table S2 in the Supplement. A total of 2 µL of the environmental DNA was added to the PCR mastermix with a total volume of 10 µL. All the reactions were set in triplicates. Quant Studio 5 was used to perform qPCR with a Power SYBR green PCR Master Mix (Invitrogen), primer concentration of 5 pM and the following amplification conditions: 95 ∘C for 10 min, 40 cycles of 95 ∘C for 15 s, 55 ∘C for 30 s and 72 ∘C for 30 s. A melting curve analysis was run after each assay to check PCR specificity. Genes encoding archaeal and bacterial 16S rRNA genes were PCR amplified from the metagenome and cloned in a TA cloning vector and plasmid DNA for each, with copy numbers 102 to 1010 used as standards for quantification purposes.
2.5 16S rRNA gene amplification and sequencing
Environmental DNA was subjected to amplification of the V4 region of the 16S rRNA gene using a Veriti 96 well thermal cycler (Applied Biosystems, Foster City, CA). The V4 region of the 16S rRNA gene was amplified using a 515F′–806R′ primer (Table S2) (Bates et al., 2010) and an amplicon library was prepared and sequenced on IonS5 NGS platform (Thermo Scientific) as per the manufacturer's protocol. The sequence reads obtained were submitted to the sequence read archive (SRA) under SRA accession: SRP155468.
2.6 Bioinformatic and statistical analysis
The reads obtained were quality filtered using the Quantitative Insights Into Microbial Ecology (QIIME 1.9.1) (Caporaso et al., 2010) bioinformatics pipeline, according to which sequences with lengths outside the bounds of 210 and 310, a mean quality score below a minimum of 25 and a maximum homopolymer run exceeding a limit of 6 were filtered out. De novo-based clustering of reads to form OTU was performed using UCLUST under QIIME workflow. Sequences with greater than 97 % similarity were assigned to the same OTU. Representative reads from each OTU were assigned taxonomy using the UCLUST trained SILVA 128 database (Quast et al., 2012). OTUs which were present in the reagent control were removed from the OTU pool of the samples. Alpha diversity parameters were calculated using alpha_diversity.py under the QIIME workflow. The OTU overlap among the DF samples and OTU pool of different DFs were elucidated using InteractiVenn (Heberle et al., 2015). Core (OTUs present in all samples) and unique OTU sequences (sequences detected in only one of the samples) were analysed using the National Center for Biotechnology Information (NCBI) BLASTn nucleotide database. A phylogenetic tree was constructed by MEGA7 software using a neighbour-joining method with 1000 bootstraps (Kumar et al., 2016).
2.7 Enrichment of sulfate-reducing bacteria (SRB) from DF samples
Two DF samples (DF1 and DF3, 10 mL in a 100 mL medium) were incubated in a Postgate medium (Postgate, 1963) containing sulfate as the sole terminal electron acceptor for 6 months under anaerobic conditions at 50 ∘C for the enrichment of sulfate-reducing bacteria. Subsequently, samples were sub-cultured thrice in the same medium and the final culture obtained was used for total DNA extraction (in triplicates) using the DNeasy® PowerSoil® kit (Qiagen 12888-50). Total DNA was extracted from 10 mL of enrichment cultures. The extracted DNA from the enrichment was processed further for 16S rRNA gene amplicon sequencing and subsequent analysis as described above.
3.1 Geochemical properties of DF samples
DF samples were analysed for their major geochemical properties (Table S1). Silicon and oxygen were the major elements present in the DF (on an average 19 % and 29 % respectively) along with Ca, S, N, Al, C and Mn. These elements were found to be abundant in bentonite-based DF (Schlumberger Oilfield Glossary, 2010). All the samples (except DF1 – pH 7.3) were found to be alkaline in nature, with the maximum for DF7 (9.17). The concentration of sulfide (S2−) was found to be in the range of 0.3–0.8 mg kg−1. High levels of barium were detected in all the DFs (except DF1) with the maximum in DF4 (46 %). With increasing sample depth, the change of pH to alkalinity reduced relative abundance of Si and Al, but enrichment of Fe, Ca and was noted (Fig. 1). A shift in geochemical parameters suggested the gradual infusion of elements from the crystalline bedrock during the course of drilling.
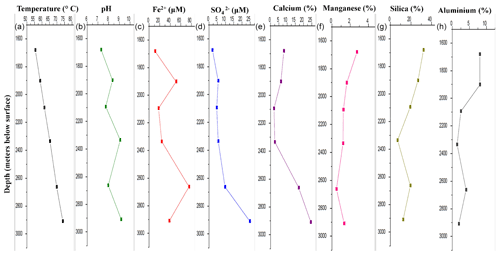
Figure 1Major geochemical parameters of DF samples. There was a distinct variation in the geochemistry of DF samples obtained at different depths. Increase in temperature (a), alkalinity (b), iron (c), sulfate (d) and calcium (e) was observed whereas the concentration of manganese (f), silica (g) and aluminium (h) showed a decreasing trend.
3.2 Bacterial and archaeal abundance in DF
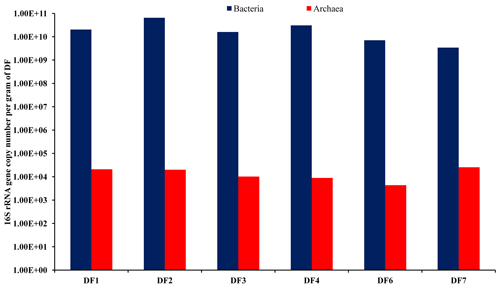
The abundance of bacterial and archaeal populations was determined by estimating the copy number of bacterial- and archaeal-specific 16S rRNA gene using a real-time PCR-based quantitative approach. With respect to the sampling depth, DF used for drilling rocks of a deeper horizon showed marginal change in bacterial (2.02×1010–3.43×109 g−1 of DF) and archaeal (2.54×104 to 4.42×103 g−1 of DF) gene copy numbers (Fig. 2). Assuming an average of 4.7 and 1.7 16S rRNA gene copies per genome of bacteria and archaea (Stoddard et al., 2015), we could estimate bacterial cell abundance ranging between 1.38×1010 and 7.29×108 g−1 of DF and archaeal cell abundance ranging between 1.22×104 and 2.55×103 g−1 of DF.
3.3 Sequencing data
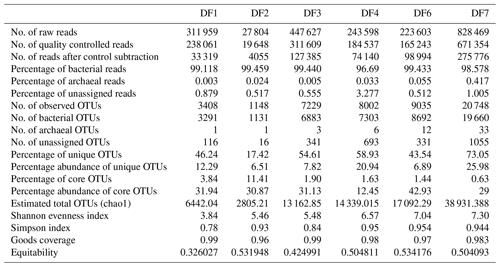
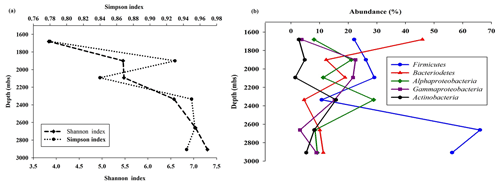
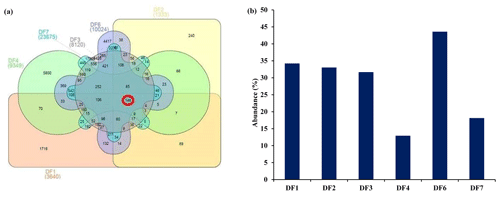
Sequencing of a hypervariable V4 region of 16S rRNA gene generated 20 83 060 reads from 6 samples (Table 1). On average 613 669 reads were found usable and these obtained reads were grouped into 49 570 operational taxonomic units (OTUs). A total of 46 950 OTUs belonged to bacteria and 56 OTUs belonged to archaea. A small fraction of OTUs (0.03 %–2.12 %) remained unassigned across all the samples. A considerable portion of OTUs (17 % to 73 %) were found to be unique and these unique OTUs represented 6 %–26 % of the total microbial community across all the samples. A total of 148 OTUs (0.29 %) were found to be common among all samples, which comprised about 25 % of the total microbial abundance. Alpha diversity analysis was performed for the sample set. Chao 1 estimator analysis suggested that samples from the deepest location (DF7) contained the highest number of OTUs. The Shannon index was in the range of 3.84–7.3 across all samples. The Simpson index ranged between 0.78 and 0.95. It could be noted that the lowest values of the Shannon and Simpson indices were obtained for DF1 and in general both the values showed an increasing trend from DF1 to DF7 (Fig. 3a).
3.4 Microbiological assessment of DF
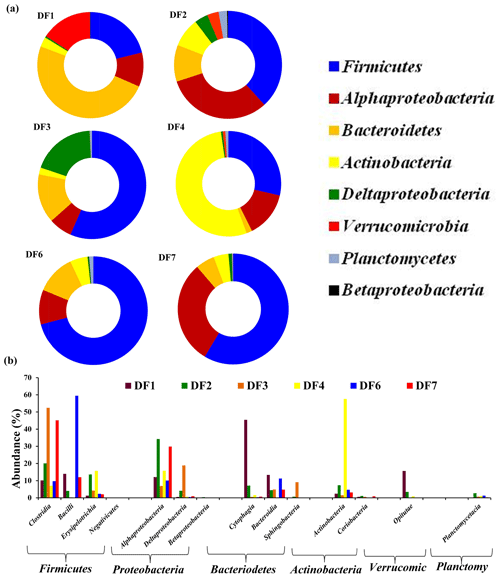
The number of sequences assigned to bacteria covered 35 phyla, 87 classes, 174 orders, 350 families and 848 genera. Sequences assigned to archaea covered 2 phyla, 5 classes, 6 orders, 6 families and 7 genera (Table 2). Based on average abundance at phylum level (phylum Proteobacteria has been further divided into its different classes), Firmicutes (10 %–66 %), Bacteriodetes (4 %–45 %), Alphaproteobacteria, (10 %) Gammaproteobacteria, (3 %–21 %) and Actinobacteria (2 %–15 %) constituted more than 90 % of the communities in the DF (Fig. 4a). Firmicutes was the most abundant phylum across all the samples except DF4. Figure 3b revealed changes in the abundance of these phyla depending on the depth. With respect to depth, the abundance of Firmicutes (20 % to 60 %), Gammaproteobacteria (3 % to 21 %) and Actinobacteria (2 % to 15 %) increased, whereas Bacteriodetes (45 % to 10 %) showed a decreasing trend. The abundance of Alphaproteobacteria remained nearly consistent across the depths (10 %–30 %). Verrucomicrobia, Planctomycetes, Spirochaetae, Tenericutes, Chloroflexi and Deinococcus-Thermus (0.07 %–3 %) were some of the other phyla observed in the DF samples. Only two archaeal phyla, namely (Fig. 4b) Euryarchaeota and Thaumarchaeota, were detected in DF samples, with relatively low abundance (0.01 %–0.4 %).

Figure 4(a) Bacterial community composition of DF samples (original and returned DF samples) at phylum level (phylum Proteobacteria has been further divided into its different classes). Phyla which had minimum 1 % abundance in the DF samples were selected and the remaining ones were grouped into “Others”. (b) Abundance of archaeal communities in DFs (phylum level).
Microbial diversity of the in-flow drilling fluid was assessed and compared with that of returning DFs (DF1–DF7). Bacteriodetes (45 %) was the most abundant phyla in the in-flow DF, followed by Firmicutes (23 %), Alphaproteobacteria and Deltaproteobacteria (Fig. 4a). These four phyla constituted more than 90 % of the total community in the in-flow fluid sample. A distinct shift in microbial community composition was observed in the returning DFs (DF1–DF7). Bacteriodetes was identified as one of the major phyla present in DF1 (44 %), and its diversity decreased in DF samples retrieved from increasing depths. Similarly, abundance of Firmicutes, Alphaproteobacteria and Gammaproteobacteria increased in the DF samples collected at lower depths. Interestingly, no archaeal phylum was detected in the in-flow sample but they were detected in the DF samples collected from the lower depths.
Clostridia, Alphaproteobacteria, Gammaproteobacteria, Bacilli, Cytophagia Actinobacteria, Bacteroidia and Betaproteobacteria (>80 %) were the major bacterial classes in DFs. Noticeably, there was an increase in the abundance of Clostridia and Bacilli in the DF samples obtained from lower depths. DF1 was majorly dominated by Cytophagia (38 %) followed by Betaproteobacteria (15 %), Clostridia (14 %), Alphaproteobacteria (8 %) and Bacteroidia (6 %). DF2 was comprised of Gammaproteobacteria (22 %) and Alphaproteobacteria (21 %), with Clostridia (11 %) being the third most abundant class. Clostridia (26 %) was the most abundant class in DF3 followed by Gamma- (21 %), Alphaproteobacteria (11 %) and Deltaproteobacteria (9 %). DF4 was majorly dominated by Alphaproteobacteria (28 %) and Gammaproteobacteria (15 %). DF4 also had a higher abundance of Actinobacteria (15 %) than other DFs. DF6 had a high abundance of Clostridia (31.5 %) and Bacilli (26.5 %) with Alphaproteobacteria (8.5 %), Actinobacteria (6 %), Bacteroidia (6 %) and Gammaproteobacteria (8 %) being the other prominent classes. DF7 had a significant abundance of Clostridia (36 %) and Bacilli (18 %) followed by Alphaproteobacteria (9 %) and Actinobacteria (5 %). Among archaeal classes Methanobacteria (DF6 and DF7) and Methanomicrobia (DF3–DF7) showed the highest abundance followed by Thermoplasmata (DF3 and DF7) and the Soil Crenarchaeotic Group (SCG) (DF6 and DF7).
3.5 Abundance of major microbial families and genera in DF
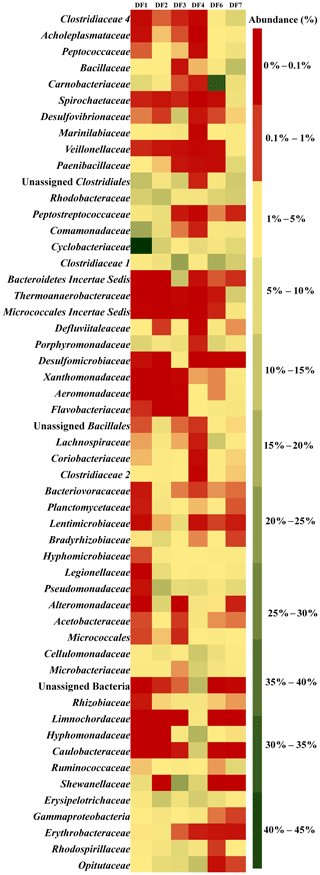
Figure 5 displays the abundance of different microbial families in DFs. DF1 was dominated by Cyclobacteriaceae (Mongoliitalea, Cecembia) (39 %), Comamonadaceae (Hydrogenophaga) (14 %) and Clostridiales (unclassified Clostridium) (9 %). Pseudomonadaceae (Pseudomaonas) (11 %), Rhodobacteraceae (Rhodobacter, Pannonibacter, Paracoccus) (10 %), Erysipelotrichaceae (Erysipelothrix) (8 %), Cyclobacteriaceae (Mongoliitalea, Cecembia) (6 %) and Alteromonadaceae (Alishewanella) (5 %) were the top five families in DF2. Shewanellaceae (Shewanella) (16 %), Clostridiaceae 1 (unclassified Clostridiaceae) (15 %), Bacteroidetes Incertae Sedis (8 %) and Desulfovibrionaceae (Desulfovibrio) (7 %) were the major families in DF3. The prominent microbial families in DF4 were Hyphomonadaceae (Hyphomonas) (12 %), Bacteria (10 %), Opitutaceae (Opitutus) (9 %), Cellulomonadaceae (Actinotalea) (8 %) and Caulobacteraceae (Brevundimonas) (6 %). Carnobacteriaceae (Marinilactibacillus, Alkalibacterium) (32 %) was found to be the most abundant microbial family in DF6 followed by Clostridiaceae 1 (Clostridium sensu stricto 1) (13 %) and Lachnospiraceae (unclassified Lachnospiraceae) (7 %). Bacillaceae (Geobacillus, Anoxybacillus, Anaerobacillus, Aeribacillus) (9 %), Clostridiaceae 1 (Clostridium sensu stricto 1) (7 %), Rhodobacteraceae (Pannonibacter) (7 %) and Thermoanaerobacteraceae (Thermoanaerobacter, Gelria) (6 %) were the major microbial families present in DF7.
Sample-wise community profiling at lower taxonomic levels highlighted the distinct microbial assemblages with change in depth. Overall community profile highlighted that with changes in depth, the DF microbial communities underwent a pronounced shift. However, members of a few bacterial taxa remained ubiquitously abundant across all the samples. Clostridiaceae, Marinilibiaceae, Rhodobactereaceae and Erysipelotrichaceae were ubiquitous in all the samples. Samples from deeper horizon (2662–2908 m b.s.) showed a distinct abundance of Clostridiaceae 4, Desulfovibrionaceae, Veillonellaceae, Aeromonadaceae and Xanthomonadaceae. On the other hand, samples recovered from shallow levels (1681–1801 m b.s.) (like DF1, DF2) were more populated with Peptococcaceae and Opitutaceae. Members of Shewanellaceae, Hyphomonadaceae and Microbacteriaceae were relatively more abundant in samples recovered from DF3 and DF4, which represent a medium depth range (2093–2335 m b.s.).
3.6 Core OTU analysis
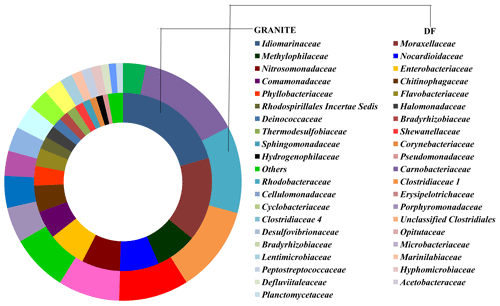
OTUs common (core OTUs) across all six DF samples were analysed. Out of the total 49 570 OTUs present across these six samples only 148 OTUs were identified as core OTUs (Fig. 6a). All of these OTUs together could cover 30 %–45 % of the total communities (Fig. 6b), with the exception of DF4 (12 %) and DF7(18 %). The relative abundance of different taxonomic groups (Fig. 7a) representing the core OTUs indicated that the abundance of Firmicutes remained higher in samples from deeper horizons (highest in DF6; 71 %). OTUs affiliated with Alphaproteobacteria represented 7 % to 31 % from DF1 to DF7. Members of Bacteriodetes present in the core OTUs represented 50 % of total communities of DF1 and 11 % to 15 % in DF2, DF3 and DF6 but reduced substantially to 2 % and 6 % in DF4 and DF7 respectively. Actinobacterial core OTUs in general constituted 2 % to 9 % of the communities, except for DF4 where these OTUs represented 53 % of total OTUs. Core OTUs affiliated with Deltaproteobacteria represented a relatively lower proportion of most of the communities except DF2 (4 %) and DF3 (19 %). Members of Verrucomicrobia, present as core OTUs, constituted 16 % of DF1 and 3 % of DF2 communities, while in the rest of the communities their abundance was very low. Planctomycetes members were generally less abundant except in DF2 (3 %). Betaproteobacterial core OTUs were no very abundant (0.004 % to 0.03 %) except in DF1 (0.13 %) and DF2 (0.29 %). Archaeal OTUs were not detected in the core communities of DF. Clostridia (24 %) and Alphaproteobacteria (18 %) were the major classes detected in the core OTUs of DF followed by Bacilli (14 %), Actinobacteria (12 %) and Cytophagia (9 %) (Fig. 7b). However, the absence of Gammaproteobacteria and lower abundance of Betaproteobacteria was noteworthy in the core OTUs of DF. The unique presence of Negativicutes and Coriobacteria (with very low abundance) was also noted within the DF core communities (Fig. 7b). Erysipelotrichia, Clostridia, Alphaproteobacteria and Deltaproteobacteria (Fig. 7b) were the other major microbial classes. Carnobacteriaceae, Rhodobacteraceae, Clostridiaceae 1, Cellulomonadaceae, Erysipelotrichaceae, Cyclobacteriaceae, Opitutaceae, Bradyrhizobiaceae and Desulfovibrionaceae were the major families present as the core community members of DF (Fig. 8). These microbial families constituting the core OTUs of DF obtained from 1681 to 2908 m b.s. were quite different from the microbial families detected in the core OTUs of granitic bedrock at higher depths (up to 1500 m b.s.). Idiomarinaceae, Moraxellaceae, Methylophilaceae, Nocardioidaceae, Nitrosomonadaceae, Enterobacteriaceae, Comamonadaceae and Chitinophagaceae were the major microbial families detected in the core OTUs of granitic bedrock up to 1500 m b.s. (Dutta et al., 2018). Marinilactibacillus, Youngilibacter, Clostridium sensu stricto 8, Geosporobacter and Pannonibacter were the highly abundant bacterial genera present in the core communities of DF.
3.7 Phylogeny of the core OTUs of DF samples
The phylogeny of the top 20 core OTUs (relative abundance 82 %) were analysed using the NCBI BLASTn nucleotide database, and a phylogenetic tree (Fig. 9) was constructed using MEGA7 software. Phylogenetic analysis revealed that the most of the core OTUs of all six DF samples displayed a significant match with known (cultured and uncultured) anaerobic, thermo-alkaliphilic microorganisms obtained from extremophilic habitats and the deep subsurface. Amphibacillus, Marinilactibacillus, Pannonibacter phragmitetus, Thermotalea metalivorans, Crassaminicella profunda, Geosporobacter subterraneus, Anaerobranca horikoshii, Anaerobranca californiensis, Crassaminicella profunda and Mongoliitalea lutea were some of the major bacteria which displayed close lineage with the OTUs of DF samples. Some of the OTUs also showed a match with uncultured thermophilic, anaerobic iron and sulfate and nitrate reducers (Clostridia, Geosporobacter, Desulfovibrio etc.).
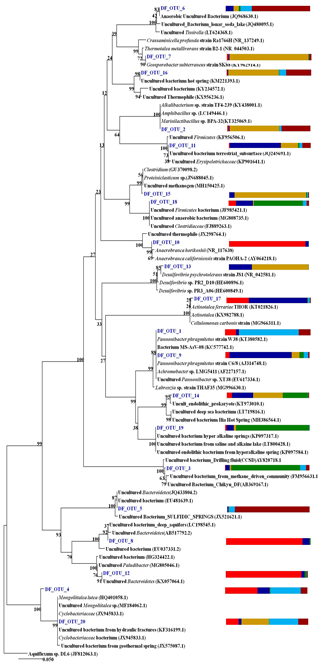
Figure 9The phylogeny of the top 20 core OTUs (relative abundance >82 %) constructed in MEGA7. Coloured stack bar represent their relative abundance in all the DF samples. Each sample is represented by different colours as follows: DF1: red, DF2: blue, DF3: yellow, DF4: green, DF6: light blue, DF7: dark red.
3.8 Phylogeny of the unique OTUs of DF samples
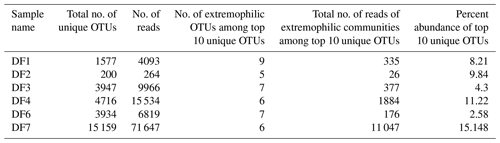
The top 10 OTUs (on the basis of relative abundance), unique to each specific DF sample, were subjected to phylogenetic analysis to investigate their identity with extremophilic lineages or microorganisms reported from the deep subsurface (Fig. 10). The relative abundance of the unique OTUs (Table 2), which showed matches with extremophiles or microorganisms reported from deep subsurface microbiome, pertaining to a specific sample, was highest for DF7 (15.42 %) and lowest for DF6 (2.58 %). Unique OTUs corresponding to DF1 showed close NCBI BLASTn matches with Hydrogenophaga aquatica, Tepidicella and other uncultured bacteria from extreme environments (hypersaline lakes) and deep environments. The top 10 unique OTUs corresponding to DF2 showed matches with Peptostreptococcaceae and Sphingobacteriales. Here also some of the OTUs showed matches with Pseudomonas strains derived from surface environments. Apart from a few OTUs pertaining to DF3 (which showed a match with bacteria from waste water and sewage sludge) most of them displayed close similarity to Desulfomicrobium and some uncultured bacterial population from deep subsurface environments. Most of the top 10 OTUs of DF4 showed correspondence to Aishwanella and other haloalkaliphilic bacteria, whereas few OTUs were related to bacteria derived from hydrocarbon-contaminated soil and anaerobic sludge. The unique OTUs (top 10) of sample DF6 exhibited close lineage with Marinilactibacillus piezotollerans and other representatives of uncultured bacteria from deep subsurface rocks and geologically derived fluids. Subsequently most of the top 10 unique OTUs of DF7 were similar to Thermincola, Thermosinus, extremotolerant Paenibacillus and other representatives from geothermal deep aquifers.
3.9 Enrichment of SRB from DF samples
16S rRNA gene amplicon sequencing and subsequent analysis revealed an assessable shift in microbial community composition of DF samples (DF1 and DF3) following anaerobic incubation under sulfate-reducing conditions. A comparison with the microbial community composition of DFs before and after enrichment is given in Fig. S1 in the Supplement. Following incubation under sulfate-reducing conditions a distinct shift in community composition with great enhancement of Firmicutes (60 %) was observed. Bacteriodetes, Alphaproteobacteria and Betaproteobacteria, which dominated the DF1 sample, decreased significantly in the enrichments. Members of the phyla Gammaproteobacteria (12 %) and Deltaproteobacteria (4 %) increased in DF1 SRB enrichments. Similarly, in the DF3 sample Firmicutes (29 %), Bacteriodetes (18 %), Gammaproteobacteria (21 %) and Alphaproteobacteria (11 %) were the major phyla. Following enrichment under sulfate-reducing conditions, a sharp increase in the abundance of Firmicutes (98 %) was noted (Fig. S1a). A 3- to 4-fold enhancement of Firmicutes was observed in the SRB enrichments for both the DF samples. Analysis of genus-level data (Fig. S1b) showed that known anaerobic sulfate reducers such as Anaerobranca, Anoxybacillus, Bacillus and Clostridium sensu stricto 8 were most prevalent in the SRB enrichments for both the DFs.
Rotary drilling has been one of the most important techniques to get the crystalline rock samples and geological fluids from deeper depths (Keift et al., 2007). The returned drilling fluid obtained on site, during the drilling of Koyna pilot borehole (at the Deccan Traps), India, contained chemical signatures of bentonite-based drilling mud (Table S1). These drilling fluids were mostly silica-rich, with Na, Al, N and C being the other major elements, and also contain a variety of organic constituents (Schlumberger Oilfield Glossary, 2010; Keift et al., 2007; Rabia, 1985). The presence of compounds such as calcium (Ca), manganese (Mn2+), sulfur (S), iron (Fe2+), sulfate () and minor amounts of sulfide (S2−) in DF samples was quite noteworthy, and their concentrations increased with increased sampling depth. Normally, those compounds should not exist in the bentonite-based DF (Montmorillonite – Handbook of Mineralogy, 2000; http://rruff.geo.arizona.edu/doclib/hom/montmorillonite.pdf, last access: 27 January 2020). With increasing sample depth, a change of pH to alkalinity and a reduced relative abundance of Si and Al was noted in the samples (Fig. 1). The granitic subsurface of the Koyna–Warna region was reported to be sulfur- and iron-rich (Misra et al., 2017). The availability of sulfate in deeper horizons (>1000 m) was widely reported through geochemical analysis of deep granitic aquifers around the globe (Ino et al., 2017, and references within). Compared to other metallic elements present in mineral form, and Fe2+ present in rock were able to dissolve in the DF during drilling; hence it got enriched with sulfate with an increase in depth. The shift in geochemical parameters suggested gradual infusion of elements from the crystalline bedrock during the course of drilling. More evidence of this infusion came from the alkaline nature of the DF samples obtained from lower depths. These granites have been reported to contain minerals responsible for creating alkaline conditions in those environments (Misra et al., 2017). The mixing of the granitic bedrock with the drilling fluid had provided alkalinity to the samples. There was a constant increase in temperature with increasing depth (55.6 to 73.9 ∘C) from 1681 to 2908 m b.s. In the subsurface of the Deccan Traps temperature increased with depth at a rate of ∼15 ∘C km−1 in granitic bedrock and lithostatic pressure increased by 26.7 MPa km−1 (Roy and Rao, 1999). The initial fluid used might have contained surface-derived microbial populations but the deep extreme subsurface environments, through which it had circulated, could have tended to favour indigenous, subsurface-adapted microorganisms (Zhang et al., 2006; Masui et al., 2008). It could also contain the microbial diversity of rock chips and geological fluids from the deep subsurface carried along with the DFs to the surface during drilling. The bacterial and archaeal cell abundance were detected using qPCR. No significant change in bacterial and archaeal abundance was observed with respect to sampling depth (Fig. 2). Microbial load in the DF was observed to be persistent across depths. Hence, it could be assumed that the change in biomass and its decay have been at equilibrium across depths in DF samples. Dynamic conditions encountered by the DF (due to drilling and coring process) might also have favoured the growth of some of the microbial populations, whereas others were not able to sustain themselves, thereby keeping the overall microbial load consistent throughout.
The microbial diversity of DFs were studied by amplicon sequencing of the V4 region of the 16S rRNA gene. This primer set (515F–806R) to amplify this region was developed by Bates et al., 2011, and was designed to be universal for nearly all bacterial and archaeal taxa. They even demonstrated in silico that this primer set could amplify 16S rRNA genes from a broad range of archaeal and bacterial groups with few biases or excluded taxa. In spite of the short-read lengths (∼250 bp), this region provided sufficient resolution for the accurate taxonomic classification of microbial sequences. Since then there have been many studies which have used the V4 region of the 16S rRNA gene to study the microbial diversity of different environments, including the deep subsurface (Dutta et al., 2018; Gupta et al., 2018; Purkamo et al., 2017). Considerably higher diversity and bacterial abundance was observed in DF with an increase in depth (Table 1). There was a marginal increase in archaeal abundance in DF with an increase in depth. Increases in Shannon and Simpson index values (Fig. 3a) confirmed the increase in the microbial diversity of DF as we went further down (Simpson, 1949; Pielou, 1966). Continuous exposure to the deep subsurface environment and its constant mixing with the granitic subsurface rocks might have enriched the microbial diversity of DF with an increase in depth (Struchtemeyer et al., 2011; Zhang et al., 2006). The phylum-level composition of microbial communities of the DF samples was similar to some of the earlier studies on the microbial diversity of DFs (Fig. 4a and b). Struchtemeyer et al., 2011, reported the dominance of Firmicutes (average 55 %) in all the DF samples from thermogenic natural gas wells of the Barnett Shale. Firmicutes were found to be the major microbial community in six DF samples collected from 2290 to 3350 m b.s. in the Chinese continental scientific drilling (CCSD) project (Zhang et al., 2006). The original in-flow DF which was collected from the tank before its introduction in the borehole had different microbial community composition than the returned DF samples (DF1–DF7) from each depth (Fig. 4a). Bacteriodetes, which dominated the in-flow DF, decreased considerably in the returned DF samples (DF1 to DF7). The phylum Bacteriodetes is widely distributed in the environment, including in soil, sediments and sea water, as well as in the guts and on the skin of animals; hence it might be indigenous to drilling fluid (Gibino et al., 2018; Rainey and Oren, 2011). Similarly, Firmicutes, Alphaproteobacteria, Actinobacteria and Gammaproteobacteria were increased in the returned DF samples. Hence, it can be seen that indigenous microbial community composition of in-flow DF was altered considerably during its interaction with the granitic rock cores during drilling. It was evident that microbial communities of the returned DF samples also varied with increases in depth due its extended exposure to deep subsurface conditions and interaction with granitic rocks. The relative abundance of some bacterial phyla such as Bacteriodetes, Betaproteobacteria and Gammaproteobacteria decreased and the relative abundance of some bacterial phyla such as Firmicutes, Alphaproteobacteria and Actinobacteria (relevant to deep subsurface) increased (Fig. 3b). There are several reports that confirmed the dominance of these phyla in the deep subsurface (Sahl et al., 2008; Leandro et al., 2018; Ino et al., 2017; Purkamo et al., 2017; Leblanc et al., 2019). The concentration of some archaeal phyla (Euryarchaeota and Thaumarchaeota) also increased in the lower depths. Interestingly, archaea were not detected in the in-flow DF. These archaeal phyla have been predominantly reported in deep igneous rocks (Nyyssönen et al., 2014; Breuker et al., 2011; Labonté et al., 2017). A dynamic shift in microbial populations was particularly noteworthy in the DF samples from the deep granitic subsurface of the Koyna–Warna region. These microbial communities must have intruded from the deep granitic rocks into the DFs during the drilling process.
The family- and genus-level data gave us better insight into the changing microbial dynamics of returning DF samples by depth (Fig. 5). Comamonadaceae and Pseudomonadaceae were observed to be dominant in DF1 (1500 m b.s.) and DF2 (1901 m b.s.) and their abundance decreased in the lower depths. Several microorganisms belonging to Commamonadaceae and Pseudomonadaceae families were mesophilic in nature, mostly found in soil and water (Palleroni, 1981; Moore et al., 2006; Willems, 2014), and they might have come either from DF or the surface. Cyclobacteriaceae and Rhodobacteriaceaeae, though dominant in DF1 and DF2, were also present in all the DF samples. These two microbial families, found in diverse and extreme habitats (freshwater bodies, algal mats, marine waters, alkaline soda lakes, hot springs, mud volcanoes etc.), manifested the ability to degrade a number of polysaccharides and have been deeply involved in sulfur and carbon biogeochemical cycling (Pujalte et al., 2014; Pinnaka and Tanuku, 2014). Major populations of these two families in DF1 and DF2 could be indigenous to DFs and surface soil or water. There is a reasonable chance that the extremophilic microbial population belonging to Cyclobacteriaceae and Rhodobacteriaceaeae might have become enriched in the DFs sampled at lower depths due to the extreme conditions (Zhang et al., 2006; Kumar et al., 2012). Alternatively, it could also be concluded that they might be part of the deep subsurface microbiome (Russell et al., 2016; Hubalek et al., 2016; Leboulanger et al., 2017). Subsequently the relative abundance of some of the microbial families, known to strive in strictly anaerobic and extremophilic environments (e.g. Thermoanaerobacteraceae, Clostridiaceae, Bacillaceae, Carnobacteriaceae, Ruminococcaceae) (Tomás et al., 2013; Deep et al., 2013; Glaring et al., 2015; Toffin et al., 2005; Ishikawa et al., 2009; Song and Dong, 2009; Horino et al., 2014), increased in the DFs sampled from 2000 to 3000 m b.s. Some of these microbial families such as Thermoanaerobacteraceae and Bacillaceae have been reported from deep subsurface environments as well as DFs from other deep subsurface studies (Chakraborty et al., 2018; Gaboyer et al., 2015; Zhang et al., 2006; Slobodkina et al., 2012; Fang et al., 2017) These groups might have intruded the DFs from the rocks at lower depths during drilling operations (Struchtemeyer et al., 2011). High concentrations of sulfate and iron in the DFs from lower depths are correlated with the presence of these microbial families which have been reported previously to utilize sulfate and iron as terminal electron acceptors (Sylvan et al., 2015; Reyes et al., 2017; Kjeldsen et al., 2007; Kanso et al., 2002; Emmerich et al., 2012; Ogg et al., 2010). The presence of thermophilic, alkaliphilic, halophilic and anaerobic fermentative bacterial genera relevant to the deep subsurface (Opitutus, Mongoliitalea, Hydrogenophaga, Marinilactibacillus, Anoxybacillus, Symbiobacterium, Geosporobacter, Thermoanaerobacter etc.) in the DFs (Lee et al., 2012; Willems et al., 1989; Toffin et al., 2005; Ueda et al., 2004; Klouche et al., 2007; Hong et al., 2015) and their dominating presence in the samples from lower depths (2500 m b.s.) confirmed the hypothesis of a possible interaction between the granitic bedrock and DF and was indicative of the possible alteration in the DF microbial community due to its exposure to the highly extreme conditions at these depths.
Core OTU analysis revealed similar observations to those stated above (Figs. 6 and 7). Some of the microbial phyla (Cyanobacteria, Chloroflexi, Chlamydiae and Euryarchaeota) indigenous to deep subsurface environments (Puente-Sánchez et al., 2014, 2018; Fullerton and Moyer, 2016; Rempfert et al., 2017) were only observed in the core OTUs of lower depths (DF4 onwards). The microbial families (Carnobacteriaceae, Rhodobacteraceae, Clostridiaceae 1, Cellulomonadaceae, Erysipelotrichaceae, Cyclobacteriaceae, Opitutaceae, Bradyrhizobiaceae and Desulfovibrionaceae) detected in the core OTUs of DF were different from the microbial families (Idiomarinaceae, Moraxellaceae, Methylophilaceae, Nocardioidaceae, Nitrosomonadaceae, Enterobacteriaceae, Comamonadaceae, Chitinophagaceae) detected in granitic core OTUs at higher depths (1500 m b.s.) (Dutta et al., 2018) (Fig. 8). These microbial families might have either come from the intrusions of subsurface deep granitic bedrock at lower depths (1600 m b.s. onwards) or might have got enriched due to the extreme environmental conditions at the deep granitic subsurface. Phylogenetic analysis of the core OTUs revealed that most of the sequences belonged to anaerobic, deep subsurface microbial communities (Fig. 9). Core OTUs displayed a good match with extremophilic microorganisms (cultured and uncultured), namely Amphibacillus (anaerobic, alkaline), Marinilactibacillus (halo-alkaliphilic, peizotolerant), Pannonibacter phragmitetus (alkalitolerant), Thermotalea metalivorans (anaerobic,thermophilic), Crassaminicella profunda (anaerobic, chemo-organotrophic bacterium), Geosporobacter subterraneus (obligate anaerobe), Anaerobranca horikoshii (alkalitolerant thermophile), Anaerobranca californiensis (obligately anaerobic, alkalithermophilic, chemo-organotroph), Crassaminicella profunda (anaerobic, chemo-organotrophic) and Mongoliitalea lutea (alkaliphilic, halotolerant). These organisms have been reported from deep subsurface environments and extreme habitats such as alkaline soda lakes, hot and alkaline springs, sulfur-containing springs, methane bedrock, deep sea sediments, and aquifers (Engle et al., 1995; Borsodi et al., 2003; Gorlenko et al., 2004; Toffin et al., 2005; Klouche et al., 2007; Wu et al., 2010; Yang et al., 2012; Lakhal et al., 2015; Ogg and Patel, 2009). Very few OTUs showed a match with both types of habitats, namely moderate as well as extreme. Phylogenetic analysis of unique OTUs pertaining to each DF sample revealed similar results (Fig. 10). Some of the OTUs unique to each sample (3 %–15 % relative abundance per sample) showed relatedness to extremophilic as well as deep subsurface microbial groups while few OTUs showed a match with mesophilic bacteria reported from surface environments. Unique OTUs corresponding to the DF1 sample showed close NCBI BLASTn matches with Hydrogenophaga aquatica, which is isolated from the hot spring (Lin et al., 2017) and Tepidicella (alkaliphilic bacteria isolated from hot spring runoff) (França et al., 2006) and other uncultured bacteria from hyperalkaline saline lakes and deep basaltic aquifers. The top 10 unique OTUs corresponding to DF2 showed matches with Peptostreptococcaceae (a moderately thermophilic anaerobic bacterial family which consists of bacteria reported from pristine rock cores, deep-sea hydrothermal vents and other extreme environments) (Slobodkin, 2014) and Sphingobacteriales (reported from deep rocks) (Dutta et al., 2018). They are similar to various Pseudomonas strains reported from river and petroleum muck environment and other uncultured bacteria from mud microbiomes as well. Uncultured bacteria from the Oukilito borehole, geothermal deep aquifer, Atlantic coast sediment (Desulfomicrobium) (Dias et al., 2008) and hydraulic fracture fluids showed close lineage with most of the top 10 unique OTUs pertaining to sample DF3, whereas a few uncultured bacteria from waste water and sewage sludge (Porphyromonadaceae) (Sakamoto, 2014) also showed relatedness to some of the unique OTUs of this sample. Some unique OTUs among the top 10 of the DF4 sample showed correlation with Alishawanella (Salah et al., 2016) and other halo-alkalophilic bacteria and were representative of hypersaline lake and deep subsurface clay rocks, and some unique OTUs were related to hydrocarbon-contaminated soil and anaerobic sludge. Unique OTUs (top 10) of sample DF6 exhibited close lineage with Marinilactibacillus piezotollerans, which is a known peizotolerant bacterium, and other representatives from hydraulic fracture fluids, deep bedrock aquifer and subsurface aquifer sediments (Toffin et al., 2005). Subsequently most of the top 10 unique OTUs of DF7 were similar to Thermincola (anaerobic, thermophilic, chemolithotrophic organism), Thermosinus (anaerobic, thermophilic and carbon monoxide oxidizing bacterium), extremotolerant Paenibacillus and other representatives from geothermal deep aquifers (Zavarina et al., 2007; Sokolova et al., 2004). Based on these observations we hypothesized that due to the prolonged interaction of DF with the subsurface granitic crust these organisms might have become infused in the DF. The interaction between DF and crust has altered the geochemistry of DF, which is evident from the altered geochemical properties of the DF. This interaction (infusion of crustal particle carrying cells) led to the enrichment of DF with extremophilic microbial populations pertaining to the deep granitic subsurface (represented by the core OTUs). On the contrary in the OTUs unique to each DF, a mixed bacterial population (both extremophilic as well as mesophilic) was observed. The presence of deep subsurface and extremophilic bacterial populations in unique as well as core OTUs confirms our hypothesis of the presence of deep subsurface microbial populations in DF samples collected at different depths below the surface. Apart from that, the presence of diverse heterotrophic bacterial populations in DF samples is quite interesting, and during core OTU analysis we observed that many OTUs showed lineages with heterotrophic bacteria. As already discussed above, DF consists of a variety of organic carbon compounds (Keift et al., 2007; Rabia, 1985). These organic carbon compounds could be efficiently utilized by heterotrophic bacteria as a carbon source and hence played an active role in shaping up the functional diversity of the DF microbial communities. Incubation of DF samples (DF1 and DF3) under sulfate-reducing conditions was quite successful and it led to increase in the abundance of Firmicutes members (Anaerobranca, Clostridium sensu stricto 8, Anoxybacillus, Bacillus) capable of anaerobic sulfate reduction (Fig. S1). All these members of the phylum Firmicutes were well known for their facultative to strict anaerobic metabolism and have been reported from several deep subsurface studies and extremophilic environments (Sahl et al., 2008; Purkamo et al., 2017; Leblanc et al., 2019; Gupta et al., 2018). Purkamo et al. (2017) detected Anaerobranca in their enrichment samples, which were set up to understand the response of acetate (carbon source) and sulfate (electron acceptor) in the fracture water collected at a depth of 2516 m b.s. at the Outokumpu drill site. As already shown these genera were also detected in DF samples (DF1 to DF7) and showed phylogenetic lineages with the bacteria isolated from extremophilic environments or the deep subsurface. A dynamic shift in the microbial community structure of DF with increases in depth was observed due to its exposure to deep subsurface environmental conditions.
Drilling fluid, often regarded as a source of contamination during investigations of deep subsurface microbiology, could be a vector for the sampling of geological fluids and signatures of microbial life from such environments. The geochemical investigation of DF that circulated through the deep borehole (down to 3000 m b.s.) from the granitic subsurface confirmed a possible interaction of DFs and igneous rocks. These interactions had conferred the DF with unique conditions (reducing, hot, alkaline and saline environments) that partly reflected the deep subsurface. Our investigations of the DFs suggested that although such circulating fluids could be responsible for contaminating the deep subsurface rock samples by introducing extraneous microbes, enrichment of these fluids with subsurface organisms was inevitable due to prolonged interaction with subsurface rocks. Successful identification and enrichment of anaerobic, thermophilic sulfate-reducing and deep subsurface relevant microbial populations from DF samples confirmed that DF could be used as a proxy to study the deep subsurface. Hence, the study of the microbial ecology of the DF that circulated through the deep borehole (down to 3000 m) from granitic subsurface of the Koyna–Warna region of the Deccan Traps provided a window through which to look into the broader spectrum of deep life residing within the deep crystalline bedrock of the Deccan Traps. The DF microbiome could also be used as a proxy for bedrock to isolate and characterize deep biosphere related microorganisms.
The sequence reads obtained were submitted to the sequence read archive (SRA) under SRA accession: SRP155468. It can be accessed from the following link: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA482760/ (Sar, 2018).
The supplement related to this article is available online at: https://doi.org/10.5194/sd-27-1-2020-supplement.
HB performed the DNA extraction, geo-microbiological analysis, statistical analysis, data organization and analysis and prepared the manuscript. AD and AG performed the amplicon sequencing of the samples and helped in statistical analysis and bioinformatics pipeline optimization. JS performed the qPCR analysis. HB, AD, AG, AR and BM did the periodic sampling. SM assisted in phylogenetic study and data interpretation. SR organized sample collection and gave inputs about the geophysical and geochemical characterization of drilling fluid, Deccan lava flow and the Koyna–Warna seismic zone. PS and SKK conceived the study, designed experiments, compiled and analysed the data, wrote the manuscript and did overall mentoring.
The authors declare that they have no conflict of interest.
The authors are thankful to all the members of the Borehole Geophysics Research Laboratory, Karad, India, engaged in the deep drilling of the Koyna pilot borehole. We are grateful to Harsh Gupta (NGRI), Shailesh Nayak (MoES) and Brijesh Kumar Bansal (MoES) for their unstinted support and motivation towards this work. Authors acknowledge the analytical instrument facility provided by the Central Research Facility, IIT Kharagpur and School of Environmental Sciences, IIT Kharagpur, for their continuous support and assistance in geochemical analysis of the samples. This study was supported by the Ministry of Earth Sciences (MoES), Government of India (project ID: MoES/P.O.(Seismo)/1(288)/2016 dated 16 March 2017). Himadri Bose is a recipient of a National Post-Doctoral Fellowship (PDF/2017/000657) from the Department of Science and Technology, Government of India. Avishek Dutta and Jayeeta Sarkar gratefully acknowledge the fellowship provided by IIT Kharagpur. Ajoy Roy gratefully acknowledges the fellowship provided by NIT Durgapur. Abhishek Gupta is a recipient of a research fellowship from the Department of Biotechnology (DBT), Government of India. Balaram Mohapatra is a recipient of an Inspire fellowship from the Department of Science and Technology, Government of India. Sourav Mukhopadhyay gratefully acknowledges the fellowship provided by Ministry of Earth Sciences (MoES), Government of India.
This research has been supported by the Ministry of Earth Sciences, Government of India (grant no. MoES/P.O.(Seismo)/1(288)/2016); the Department of Science and Technology, Government of India (grant no. PDF/2017/000657); IIT Kharagpur; NIT Durgapur; the Department of Biotechnology (DBT), Government of India; the Department of Science and Technology, Government of India; and the Ministry of Earth Sciences (MoES), Government of India.
This paper was edited by Jan Behrmann and reviewed by two anonymous referees.
Bates, S. T., Berg-Lyons, D., Caporaso, J. G., Walters, W. A., Knight, R., and Fierer, N.: Examining the global distribution of dominant archaeal populations in soil, ISME J., 5, 908–917, https://doi.org/10.1038/ismej.2010.171, 2010.
Beeman, R. E. and Suflita, J. M.: Evaluation of deep subsurface sampling procedures using serendipitous microbial contaminats as tracers organisms, Geomicrobiol. J., 7, 223–233, https://doi.org/10.1080/01490458909377868, 1989.
Borgonie, G., Magnabosco, C., García-Moyano, A., Linage-Alvarez, B., Ojo, A. O., Freese, L. B., Van Jaarsveld, C., Van Rooyen, C., Kuloyo, O., Cason, E. D., and Vermeulen, J.: New ecosystems in the deep subsurface follow the flow of water driven by geological activity, Sci. Rep., 9, 3310, https://doi.org/10.1038/s41598-019-39699-w, 2019.
Borsodi, A. K., Micsinai, A., Kovacs, G., Toth, E., Schumann, P., Kovacs, A. L., Böddi, B., and Marialigeti, K.: Pannonibacter phragmitetus gen. nov., sp. nov., a novel alkalitolerant bacterium isolated from decomposing reed rhizomes in a Hungarian soda lake, Int. J. Syst. Evol. Microbiol., 53, 555–561, https://doi.org/10.1099/ijs.0.02356-0, 2003.
Breuker, A., Köweker, G., Blazejak, A., and Schippers, A.: The deep biosphere in terrestrial sediments in the Chesapeake Bay area, Virginia, USA, Front. Microbiol., 2, 156, https://doi.org/10.3389/fmicb.2011.00156, 2011.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., Fierer, N., Pena, A. G., Goodrich, J. K., Gordon, J. I., and Huttley, G. A.: QIIME allows analysis of high-throughput community sequencing data, Nat. Methods, 7, 335–336, 2010.
Chakraborty, A., Ellefson, E., Li, C., Gittins, D., Brooks, J. M., Bernard, B. B., and Hubert, C. R.: Thermophilic endospores associated with migrated thermogenic hydrocarbons in deep Gulf of Mexico marine sediments, ISME J., 12, 1895–1906, 2018.
Colwell, F. S. and D'Hondt, S.: Nature and extent of the deep biosphere, Rev. Mineral. Geochem., 75, 547–574, https://doi.org/10.2138/rmg.2013.75.17, 2013.
Deep, K., Poddar, A., and Das, S. K.: Anoxybacillus suryakundensis sp. nov, a moderately thermophilic, alkalitolerant bacterium isolated from hot spring at Jharkhand, India, PloS One, 8, e85493, https://doi.org/10.1371/journal.pone.0085493, 2013.
Dias, M., Salvado, J. C., Monperrus, M., Caumette, P., Amouroux, D., Duran, R., and Guyoneaud, R.: Characterization of Desulfomicrobium salsuginis sp. nov. and Desulfomicrobium aestuarii sp. nov., two new sulfate-reducing bacteria isolated from the Adour estuary (French Atlantic coast) with specific mercury methylation potentials, Syst. Appl. Microbiol., 31, 30–37, https://doi.org/10.1016/j.syapm.2007.09.002, 2008.
Dutta, A., Dutta Gupta, S., Gupta, A., Sarkar, J., Roy, S., Mukherjee, A., and Sar, P.: Exploration of deep terrestrial subsurface microbiome in Late Cretaceous Deccan traps and underlying Archean basement, India, Sci. Rep., 8, 17459, https://doi.org/10.1038/s41598-018-35940-0, 2018.
Emmerich, M., Bhansali, A., Lösekann-Behrens, T., Schröder, C., Kappler, A., and Behrens, S.: Abundance, distribution, and activity of Fe (II)-oxidizing and Fe (III)-reducing microorganisms in hypersaline sediments of Lake Kasin, southern Russia, Appl. Environ. Microbiol., 78, 4386–4399, https://doi.org/10.1128/AEM.07637-11, 2012.
Engle, M., Li, Y., Woese, C., and Wiegel, J.: Isolation and characterization of a novel alkalitolerant thermophile, Anaerobranca horikoshii gen. nov., sp. nov., Int. J. Syst. Evol. Microbiol., 45, 454–461, https://doi.org/10.1099/ijs.0.004218-0, 1995.
Escudero, C., Oggerin, M., and Amils, R.: The deep continental subsurface: The dark biosphere, Int. Microbiol., 21.3–14, https://doi.org/10.1007/s10123-018-0009-y, 2018.
Fang, J., Kato, C., Runko, G.M., Nogi, Y., Hori, T., Li, J., Morono, Y., and Inagaki, F.: Predominance of viable spore-forming piezophilic bacteria in high-pressure enrichment cultures from ∼1.5 to 2.4 km-deep coal-bearing sediments below the ocean floor, Front. Microbiol., 8, 137, https://doi.org/10.3389/fmicb.2017.00137, 2017.
Flemming, H. C. and Wuertz, S.: Bacteria and archaea on Earth and their abundance in biofilms, Nat. Rev. Microbiol., 17, 247–260, https://doi.org/10.1038/s41579-019-0158-9, 2019.
França, L., Rainey, F. A., Nobre, M. F., and Da Costa, M. S.: Tepidicella xavieri gen. nov., sp. nov., a betaproteobacterium isolated from a hot spring runoff, Int. J. Syst. Evol. Microbiol., 56, 907–912, https://doi.org/10.1099/ijs.0.64193-0, 2006.
Fredrickson, J. K. and Balkwill, D. L.: Geomicrobial processes and biodiversity in the deep terrestrial subsurface, Geomicrobiol. J., 23, 345–356, https://doi.org/10.1080/01490450600875571, 2006.
Fullerton, H. and Moyer, C. L.: Comparative single-cell genomics of Chloroflexi from the Okinawa Trough deep subsurface biosphere, Appl. Environ. Microbiol., 82, 3000–3008, https://doi.org/10.1128/AEM.00624-16, 2016.
Gaboyer, F., Burgaud, G., and Alain, K.: Physiological and evolutionary potential of microorganisms from the Canterbury Basin subseafloor, a metagenomic approach, FEMS Microbiol. Ecol., 9, 5, https://doi.org/10.1093/femsec/fiv029, 2015.
Gandhi, S. M. and Sarkar, B. C. (Eds.): Essentials of Mineral Exploration and Evaluation, Elsevier, Amesterdam, the Netherlands, 2003.
Gibiino, G., Lopetuso, L. R., Scaldaferri, F., Rizzatti, G., Binda, C., and Gasbarrini, A.: Exploring Bacteroidetes: Metabolic key points and immunological tricks of our gut commensals, Dig. Liver Dis., 50, 635–639, https://doi.org/10.1016/j.dld.2018.03.016, 2018.
Glaring, M. A., Vester, J. K., Lylloff, J. E., Al-Soud, W. A., Sørensen, S. J., and Stougaard, P.: Microbial diversity in a permanently cold and alkaline environment in Greenland, PloS One, 10, e0124863, https://doi.org/10.1371/journal.pone.0124863, 2015.
Gorlenko, V., Tsapin, A., Namsaraev, Z., Teal, T., Tourova, T., Engler, D., Mielke, R., and Nealson, K.: Anaerobranca californiensis sp. nov., an anaerobic, alkalithermophilic, fermentative bacterium isolated from a hot spring on Mono Lake, Syst. Evol. Microbiol., 54, 739–743, https://doi.org/10.1099/ijs.0.02909-0, 2004.
Guerra, A. B., Oliveira, J. S., Silva-Portela, R. C., Araujo, W., Carlos, A. C., Vasconcelos, A. T. R., Freitas, A. T., Domingos, Y. S., de Farias, M. F., Fernandes, G. J. T., and Agnez-Lima, L. F.: Metagenome enrichment approach used for selection of oil-degrading bacteria consortia for drill cutting residue bioremediation, Environ. Pollut., 235, 869–880, https://doi.org/10.1016/j.envpol.2018.01.014, 2018.
Gupta, A., Dutta, A., Sarkar, J., Panigrahi, M. K., and Sar, P.: Low-Abundance Members of the Firmicutes Facilitate Bioremediation of Soil Impacted by Highly Acidic Mine Drainage From the Malanjkhand Copper Project, India, Front. Microbiol., 9, 2882, https://doi.org/10.3389/fmicb.2018.02882, 2018.
Heberle, H., Meirelles, G. V., da Silva, F. R., Telles, G. P., and Minghim, R.: InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams, BMC Bioinformatics, 16, 169, https://doi.org/10.1186/s12859-015-0611-3, 2015.
Hoehler, T. M. and Jørgensen, B. B.: Microbial life under extreme energy limitation, Nat. Rev. Microbiol., 11, 83–94, 2013.
Hong, H., Kim, S. J., Min, U. G., Lee, Y. J., Kim, S. G., Jung, M. Y., Seo, Y. S., and Rhee, S. K.: Geosporobacter ferrireducens sp. nov., an anaerobic iron-reducing bacterium isolated from an oil-contaminated site, Antonie van Leeuwenhoek, 107, 971–977, https://doi.org/10.1007/s10482-015-0389-3, 2015.
Horino, H., Fujita, T., and Tonouchi, A.: Description of Anaerobacterium chartisolvens gen. nov., sp. nov., an obligately anaerobic bacterium from Clostridium rRNA cluster III isolated from soil of a Japanese rice field, and reclassification of Bacteroides cellulosolvens Murray et al. 1984 as Pseudobacteroides cellulosolvens gen. nov., comb. nov., Int. J. Syst. Evol. Microbiol., 64, 1296–1303, https://doi.org/10.1099/ijs.0.059378-0, 2014.
Hubalek, V., Wu, X., Eiler, A., Buck, M., Heim, C., Dopson, M., Bertilsson, S., and Ionescu, D.: Connectivity to the surface determines diversity patterns in subsurface aquifers of the Fennoscandian shield, ISME J., 10, 2556, https://doi.org/10.1038/ismej.2016.94, 2016.
Ino, K., Hernsdorf, A. W., Konno, U., Kouduka, M., Yanagawa, K., Kato, S., Sunamura, M., Hirota, A., Togo, Y. S., Ito, K., and Fukuda, A.: Ecological and genomic profiling of anaerobic methane-oxidizing archaea in a deep granitic environment, ISME J., 12, 31–47, https://doi.org/10.1038/ismej.2017.140, 2017.
İşçi, E. and Turutoğlu, S. İ.: Stabilization of the mixture of bentonite and sepiolite as a water based drilling fluid, J. Pet. Sci. Eng., 76, 1–5, https://doi.org/10.1016/j.petrol.2010.11.021, 2011.
Ishikawa, M., Tanasupawat, S., Nakajima, K., Kanamori, H., Ishizaki, S., Kodama, K., Okamoto-Kainuma, A., Koizumi, Y., Yamamoto, Y., and Yamasato, K.: Alkalibacterium thalassium sp. nov., Alkalibacterium pelagium sp. nov., Alkalibacterium putridalgicola sp. nov. and Alkalibacterium kapii sp. nov., slightly halophilic and alkaliphilic marine lactic acid bacteria isolated from marine organisms and salted foods collected in Japan and Thailand, Int. J. Syst. Evol. Microbiol., 59, 1215–1226, https://doi.org/10.1099/ijs.0.65602-0, 2009.
Islam, E., Paul, D., and Sar, P.: Microbial diversity in Uranium deposits from Jaduguda and Bagjata Uranium mines, India as revealed by clone library and denaturing gradient gel electrophoresis analyses, Geomicrobiol. J., 31, 862–874, https://doi.org/10.1080/01490451.2014.907375, 2014.
Kanso, S., Greene, A. C., and Patel, B. K.: Bacillus subterraneus sp. nov., an iron-and manganese-reducing bacterium from a deep subsurface Australian thermal aquifer, Int. J. Syst. Evol. Microbiol., 52, 869–874, https://doi.org/10.1099/00207713-52-3-869, 2002.
Kieft, T. L.: Sampling the deep sub-surface using drilling and coring techniques, in: Handbook of Hydrocarbon and Lipid Microbiology, edited by: Timmis, K. N., Springer, Berlin, Heidelberg, 3427–3441, https://doi.org/10.1007/978-3-540-77587-4_267, 2010.
Kieft, T. L.: Microbiology of the Deep Continental Biosphere, in: Their World: A Diversity of Microbial Environments, edited by: Hurst, C., Springer, Cham, 225–249, https://doi.org/10.1007/978-3-319-28071-4, 2016.
Kieft, T. L., Phelps, T. J., and Fredrickson, J. K.: Drilling, coring, and sampling subsurface environments, in: Manual of Environmental Microbiology, 3rd edition, edited by: Hurst, C. J., ASM Press, Washington, D.C., USA, 799–817, https://doi.org/10.1128/9781555815882.ch66, 2007.
Kjeldsen, K. U., Loy, A., Jakobsen, T. F., Thomsen, T. R., Wagner, M., and Ingvorsen, K.: Diversity of sulfate-reducing bacteria from an extreme hypersaline sediment, Great Salt Lake (Utah), FEMS Microbiol. Ecol., 60, 287–298, https://doi.org/10.1111/j.1574-6941.2007.00288.x, 2007.
Klouche, N., Fardeau, M. L., Lascourreges, J. F., Cayol, J. L., Hacene, H., Thomas, P., and Magot, M.: Geosporobacter subterraneus gen. nov., sp. nov., a spore-forming bacterium isolated from a deep subsurface aquifer, Int. J. Syst. Evol. Microbiol., 57, 1757–1761, https://doi.org/10.1099/ijs.0.64642-0, 2007.
Kumar, P. A., Srinivas, T. N. R., Madhu, S., Sravan, R., Singh, S., Naqvi, S. W. A., Mayilraj, S., and Shivaji, S.: Cecembia lonarensis gen. nov., sp. nov., a haloalkalitolerant bacterium of the family Cyclobacteriaceae, isolated from a haloalkaline lake and emended descriptions of the genera Indibacter, Nitritalea and Belliella, Int. J. Syst. Evol. Microbiol., 62, 2252–2258, https://doi.org/10.1099/ijs.0.038604-0, 2012.
Kumar, S., Stecher, G., and Tamura, K.: MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets, Mol. Biol Evol., 33, 1870–1874, https://doi.org/10.1093/molbev/msw054, 2016.
Labonté, J. M., Lever, M. A., Edwards, K. J., and Orcutt, B. N.: Influence of igneous basement on deep sediment microbial diversity on the eastern Juan de Fuca ridge flank, Front. Microbiol., 8, 1434, https://doi.org/10.3389/fmicb.2017.01434, 2017.
Lakhal, R., Pradel, N., Postec, A., Ollivier, B., Cayol, J. L., Godfroy, A., Fardeau, M. L., and Galés, G.: Crassaminicella profunda gen. nov., sp. nov., an anaerobic marine bacterium isolated from deep-sea sediments, Int. J. Syst. Evol. Microbiol., 65, 3097–3102, https://doi.org/10.1099/ijsem.0.000386, 2015.
Leandro, T., Rodriguez, N., Rojas, P., Sanz, J. L., da Costa, M. S., and Amils, R.: Study of methanogenic enrichment cultures of rock cores from the deep subsurface of the Iberian Pyritic Belt, Heliyon, 4, e00605, https://doi.org/10.1016/j.heliyon.2018.e00605, 2018.
Leblanc, V., Hellal, J., Fardeau, M.L., Khelaifia, S., Sergeant, C., Garrido, F., Ollivier, B., and Joulian, C.: Microbial and Geochemical Investigation down to 2000 m Deep Triassic Rock (Meuse/Haute Marne, France), Geosciences, 9, 3, https://doi.org/10.3390/geosciences9010003, 2019.
Leboulanger, C., Agogué, H., Bernard, C., Bouvy, M., Carré, C., Cellamare, M., Duval, C., Fouilland, E., Got, P., Intertaglia, L., and Lavergne, C.: Microbial diversity and cyanobacterial production in Dziani Dzaha crater lake, a unique tropical thalassohaline environment, PloS One, 12, e0168879, https://doi.org/10.1371/journal.pone.0168879, 2017.
Lee, S. J., Lee, Y. J., Ryu, N., Park, S., Jeong, H., Lee, S. J., Kim, B. C., Lee, D. W., and Lee, H. S.: Draft genome sequence of the thermophilic bacterium Anoxybacillus kamchatkensis G10, J. Bacteriol., 194, 6684–6685, https://doi.org/10.1128/JB.01877-12, 2012.
Lin, S. Y., Hameed, A., Wen, C. Z., Hsu, Y. H., Liu, Y. C., Lai, W. A., and Young, C. C.: Hydrogenophaga aquatica sp. nov., isolated from a hot spring, Int. J. Syst. Evol. Microbiol., 67, 3716–3721, https://doi.org/10.1099/ijsem.0.002146, 2017.
Liu, S. V., Zhou, J., Zhang, C., Cole, D. R., Gajdarziska-Josifovska, M., and Phelps, T. J.: Thermophilic Fe (III)-reducing bacteria from the deep subsurface: the evolutionary implications, Science, 277, 1106–1109, https://doi.org/10.1126/science.277.5329.1106, 1997.
Magnabosco, C., Lin, L. H., Dong, H., Bomberg, M., Ghiorse, W., Stan-Lotter, H., Pedersen, K., Kieft, T. L., van Heerden, E., and Onstott, T. C.: The biomass and biodiversity of the continental subsurface, Nat. Geosci., 11, 707–717, 2018.
Masui, N., Morono, Y., and Inagaki, F.: Microbiological assessment of circulation mud fluids during the first operation of riser drilling by the deep-earth research vessel Chikyu, Geomicrobiol. J., 25, 274–282, https://doi.org/10.1080/01490450802258154, 2008.
McMahon, S. and Parnell, J.: Weighing the deep continental biosphere, FEMS Microbiol. Ecol., 87, 113–120, https://doi.org/10.1111/1574-6941.12196, 2014.
McMahon, S. and Parnell, J.: The deep history of Earth's biomass, J. Geol. Soc., 175, 716–720, https://doi.org/10.1144/jgs2018-061, 2018.
Montmorillonite – Handbook of Mineralogy: Mineral data, available at: http://rruff.geo.arizona.edu/doclib/hom/montmorillonite.pdf, (last access: 28 January 2020) 2000.
Misra, S., Roy, S., Bartakke, V., Athavale, G., and Gupta, H.: Fissures and fractures in the Koyna seismogenic zone, western India, J. Geol. Soc. India, 90, 131–137, https://doi.org/10.1007/s12594-017-0690-z, 2017.
Miteva, V., Burlingame, C., Sowers, T., and Brenchley, J.: Comparative evaluation of the indigenous microbial diversity vs. drilling fluid contaminants in the NEEM Greenland ice core, FEMS Microbiol. Ecol., 89, 238–256, https://doi.org/10.1111/1574-6941.12286, 2014.
Moore, E. R., Tindall, B. J., Dos Santos, V. A. M., Pieper, D. H., Ramos, J. L., and Palleroni, N. J.: Nonmedical: pseudomonas, in: The prokaryotes, Springer, New York, NY, USA, 646–703, https://doi.org/10.1007/0-387-30746-x_21, 2006.
Nyyssönen, M., Hultman, J., Ahonen, L., Kukkonen, I., Paulin, L., Laine, P., Itävaara, M., and Auvinen, P.: Taxonomically and functionally diverse microbial communities in deep crystalline rocks of the Fennoscandian shield, ISME J., 8, 126–138, 2014.
Ogg, C. D. and Patel, B. K.: Thermotalea metallivorans gen. nov., sp. nov., a thermophilic, anaerobic bacterium from the Great Artesian Basin of Australia aquifer, Int. J. Syst. Evol. Microbiol., 59, 964–971, https://doi.org/10.1099/ijs.0.004218-0, 2009.
Ogg, C. D., Greene, A. C., and Patel, B. K.: Thermovenabulum gondwanense sp. nov., a thermophilic anaerobic Fe (III)-reducing bacterium isolated from microbial mats thriving in a Great Artesian Basin bore runoff channel, Int. J. Syst. Evol. Microbiol., 60, 1079–1084, https://doi.org/10.1099/ijs.0.009886-0, 2010.
Onstott, T. C., Phelps, T. J., Colwell, F. S., Ringelberg, D., White, D. C., and Boone, D. R.: Observations pertaining to the origin and ecology of microorganisms recovered from the deep subsurface of Taylorsville Bain, Virginia, Geomicrobiol. J., 15, 353–385, https://doi.org/10.1080/01490459809378088, 1998.
Palleroni, N. J.: Introduction to the Family Pseudomonadaceae, in: The Prokaryotes, edited by: Starr, M. P., Stolp, H., Trüper, H. G., Balows, A., and Schlegel, H. G., Springer, Berlin, Heidelberg, https://doi.org/10.1007/978-3-662-13187-9_59, 1981.
Pedersen, K., Hallbeck, L., Arlinger, J., Erlandson, A. C., and Jahromi, N.: Investigation of the potential for microbial contamination of deep granitic aquifers during drilling using 16S rRNA gene sequencing and culturing methods, J. Microbiol. Meth., 30, 179–192, https://doi.org/10.1016/S0167-7012(97)00066-3, 1997.
Pielou, E. C.: Species-diversity and pattern-diversity in the study of ecological succession, J. Theor. Biol., 10, 370–383, https://doi.org/10.1016/0022-5193(66)90133-0, 1966.
Pinnaka, A. K. and Tanuku, N. R. S.: The Family Cyclobacteriaceae, in: The Prokaryotes, edited by: Rosenberg, E., DeLong, E. F., Lory, S., Stackebrandt, E., and Thompson, F., Springer, Berlin, Heidelberg, https://doi.org/10.1007/978-3-642-38954-2_139, 2014.
Postgate, J. R.: Versatile medium for the enumeration of sulfate-reducing bacteria, Appl. Environ. Microbiol., 11, 265–267, 1963.
Puente-Sánchez, F., Moreno-Paz, M., Rivas, L.A., Cruz-Gil, P., García-Villadangos, M., Gómez, M.J., Postigo, M., Garrido, P., González-Toril, E., Briones, C., and Fernández-Remolar, D.: Deep subsurface sulfate reduction and methanogenesis in the Iberian Pyrite Belt revealed through geochemistry and molecular biomarkers, Geobiology, 12, 34–47, https://doi.org/10.1111/gbi.12065, 2014.
Puente-Sánchez, F., Arce-Rodríguez, A., Oggerin, M., García-Villadangos, M., Moreno-Paz, M., Blanco, Y., Rodríguez, N., Bird, L., Lincoln, S. A., Tornos, F., and Prieto-Ballesteros, O.: Viable cyanobacteria in the deep continental subsurface, Pr. Natl. Acad. Sci. USA, 115, 10702–10707, https://doi.org/10.1073/pnas.1808176115, 2018.
Pujalte, M. J., Lucena, T., Ruvira, M. A., Arahal, D. R., and Macián, M. C.: The Family Rhodobacteraceae, in: The Prokaryotes, edited by: Rosenberg, E., DeLong, E. F., Lory, S., Stackebrandt, E., and Thompson, F., Springer, Berlin, Heidelberg, https://doi.org/10.1007/978-3-642-30197-1_377, 2014.
Purkamo, L., Bomberg, M., Nyyssönen, M., Ahonen, L., Kukkonen, I., and Itävaara, M.: Response of deep subsurface microbial community to different carbon sources and electron acceptors during ∼2 months incubation in microcosms, Front. Microbiol., 8, 232, https://doi.org/10.3389/fmicb.2017.00232, 2017.
Purkamo, L., Kietäväinen, R., Miettinen, H., Sohlberg, E., Kukkonen, I., Itävaara, M., and Bomberg, M.: Diversity and functionality of archaeal, bacterial and fungal communities in deep Archaean bedrock groundwater, FEMS Microbiol. Ecol., 94, 8, https://doi.org/10.1093/femsec/fiy116, 2018.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glöckner, F. O.: The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res., 41, D590–D596, https://doi.org/10.1093/nar/gks1219, 2012.
Rabia, H. (Ed.): Oil Well Drilling Engineering: Principles and Practice, Graham & Trotman. Inc., London, UK, 1985.
Rainey, F. and Oren, A. (Eds.): Taxonomy of prokaryotes, Academic Press, London, UK, 2011.
Rempfert, K. R., Miller, H. M., Bompard, N., Nothaft, D., Matter, J. M., Kelemen, P., Fierer, N., and Templeton, A. S.: Geological and geochemical controls on subsurface microbial life in the Samail Ophiolite, Oman, Front. Microbiol., 8, 56, https://doi.org/10.3389/fmicb.2017.00056, 2017.
Reyes, C., Schneider, D., Thürmer, A., Kulkarni, A., Lipka, M., Sztejrenszus, S. Y., Böttcher, M. E., Daniel, R., and Friedrich, M. W.: Potentially active iron, sulfur, and sulfate reducing bacteria in Skagerrak and Bothnian Bay sediments, Geomicrobiol. J., 34, 840–850, https://doi.org/10.1080/01490451.2017.1281360, 2017.
Roh, Y., Liu, S. V., Li, G., Huang, H., Phelps, T. J., and Zhou, J.: Isolation and characterization of metal-reducing thermoanaerobacter strains from deep subsurface environments of the Piceance Basin, Colorado, Appl. Environ. Microbiol., 68, 6013–6020, https://doi.org/10.1128/AEM.68.12.6013-6020.2002, 2002.
Roy, S. and Rao, R. U. M.: Geothermal investigations in the 1993 Latur earthquake area, Deccan volcanic province, India, Tectonophysics, 306, 237–252, https://doi.org/10.1016/S0040-1951(99)00051-7, 1999.
Roy, S. and Rao, R. U. M.: Heat flow in the Indian shield, J. Geophys. Res.-Sol. Ea., 105, 25587–25604, https://doi.org/10.1029/2000JB900257, 2000.
Russell, J. A., León-Zayas, R., Wrighton, K., and Biddle, J. F.: Deep subsurface life from North Pond: enrichment, isolation, characterization and genomes of heterotrophic bacteria, Front. Microbiol., 7, 678, https://doi.org/10.3389/fmicb.2016.00678, 2016.
Sahl, J. W., Schmidt, R., Swanner, E. D., Mandernack, K. W., Templeton, A. S., Kieft, T. L., Smith, R. L., Sanford, W. E., Callaghan, R. L., Mitton, J. B., and Spear, J. R.: Subsurface microbial diversity in deep-granitic-fracture water in Colorado, Appl. Environ. Microbiol., 74, 143–152, https://doi.org/10.1128/AEM.01133-07, 2008.
Sakamoto, M.: The Family Porphyromonadaceae, in: The Prokaryotes, Springer, Berlin, Heidelberg, 811–824, https://doi.org/10.1007/978-3-642-38954-2_132, 2014.
Salah, Z. B., Rout, S. P., and Humphreys, P. N.: Draft whole-genome sequence of the alkaliphilic Alishewanella aestuarii strain HH-ZS, isolated from historical lime kiln waste-contaminated soil, Genome Announce., 4, e01447-16, https://doi.org/10.1128/genomeA.01447-16, 2016.
Sar, P.: Terrestrial deep subsurface Raw sequence reads, available at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA482760/ (last access: 10 February 2020), 2018.
Schlumberger Oilfield Glossary: Terms: Electrical Stability Test, Electrical Resitivity, Invert Emulsion, available at: http://glossary.connect.slb.com (last access: 28 January 2020), 2010.
Simpson, E. H.: Measurement of species diversity, Nature, 163, 688, https://doi.org/10.1038/163688a0, 1949.
Slobodkin, A.: The Family Peptostreptococcaceae, in: The Prokaryotes, Springer, Berlin, Heidelberg, 291–302, https://doi.org/10.1007/978-3-642-30120-9_217, 2014.
Slobodkina, G. B., Kolganova, T. V., Kostrikina, N. A., Bonch-Osmolovskaya, E. A., and Slobodkin, A. I.: Caloribacterium cisternae gen. nov., sp. nov., an anaerobic thermophilic bacterium from an underground gas storage reservoir, Int. J. Syst. Evol. Microbiol., 62, 1543–1547, https://doi.org/10.1099/ijs.0.033076-0, 2012.
Smith, D. C., Spivack, A. J., Fisk, M. R., Haveman, S. A., and Staudigel, H.: Tracer-based estimates of drilling-induced microbial contamination of deep sea crust, Geomicrobiol. J., 17, 207–219, https://doi.org/10.1080/01490450050121170, 2000.
Soares, A., Edwards, A., An, D., Bagnoud, A., Bomberg, M., Budwill, K., Caffrey, S., Fields, M., Toner, B., Gralnick, J., and Kadnikov, V.: A global perspective on microbial diversity in the terrestrial deep subsurface, bioRxiv, p602672, https://doi.org/10.1101/602672, in review, 2019.
Sokolova, T., González Grau, J. M., Kostrikina, N. A., Chernyh, N. A., Kochetkova, T. V., Bonch-Osmolovskaya, E. A., and Robb, F.: Thermosinus carboxydivorans gen. nov., sp. nov., a new anaerobic, thermophilic, carbon-monoxide-oxidizing, hydrogenogenic bacterium from a hot pool of Yellowstone National Park, Int. J. Syst. Evol. Microbiol., 54, 2353–2359, https://doi.org/10.1099/ijs.0.63186-0, 2004.
Song, L. and Dong, X.: Hydrogenoanaerobacterium saccharovorans gen. nov., sp. nov., isolated from H2-producing UASB granules, Int. J. Syst. Evol. Microbiol., 59, 295–299, https://doi.org/10.1099/ijs.0.000349-0, 2009.
Stoddard, S. F., Smith, B. J., Hein, R., Roller, B. R. K., and Schmidt, T. M.: rrn DB: Improved tools for interpreting rRNA gene abundance in bacteria and archaea and a newfoundation for future development, Nucleic Acids Res., 43, D593–D598, https://doi.org/10.1093/nar/gku1201, 2015.
Struchtemeyer, C. G., Davis, J. P., and Elshahed, M. S.: Influence of the drilling mud formulation process on the bacterial communities in thermogenic natural gas wells from the Barnett Shale, Appl. Environ. Microbiol, 7, AEM-00233, https://doi.org/10.1128/AEM.00233-11, 2011.
Sylvan, J. B., Hoffman, C. L., Momper, L. M., Toner, B. M., Amend, J. P., and Edwards, K. J.: Bacillus rigiliprofundi sp. nov., an endospore-forming, Mn-oxidizing, moderately halophilic bacterium isolated from deep subseafloor basaltic crust, Int. J. Syst. Evol. Microbiol., 65, 1992–1998, https://doi.org/10.1099/ijs.0.000211, 2015.
Toffin, L., Zink, K., Kato, C., Pignet, P., Bidault, A., Bienvenu, N., Birrien, J. L., and Prieur, D.: Marinilactibacillus piezotolerans sp. nov., a novel marine lactic acid bacterium isolated from deep sub-seafloor sediment of the Nankai Trough, Int. J. Syst. Evol. Microbiol., 55, 345–351, https://doi.org/10.1099/ijs.0.63236-0, 2005.
Tomás, A. F., Karakashev, D., and Angelidaki, I.: Thermoanaerobacter pentosaceus sp. nov., an anaerobic, extremely thermophilic, high ethanol-yielding bacterium isolated from household waste, Int. J. Syst. Evol. Microbiol., 63, 2396–2404, https://doi.org/10.1099/ijs.0.045211-0, 2013.
Ueda, K., Yamashita, A., Ishikawa, J., Shimada, M., Watsuji, T. O., Morimura, K., Ikeda, H., Hattori, M., and Beppu, T.: Genome sequence of Symbiobacterium thermophilum, an uncultivable bacterium that depends on microbial commensalism, Nucleic. Acids. Res., 32, 4937–4944, https://doi.org/10.1093/nar/gkh830, 2004.
Whitman, W. B., Coleman, D. C., and Wiebe, W. J.: Prokaryotes: the unseen majority, P. Natl. Acad. Sci. USA, 95, 6578–6583, https://doi.org/10.1073/pnas.95.12.6578, 1998.
Willems, A.: The Family Comamonadaceae, in: The Prokaryotes, edited by: Rosenberg, E., DeLong, E. F., Lory, S., Stackebrandt, E., and Thompson, F., Springer, Berlin, Heidelberg, Germany, 2014.
Willems, A., Busse, J., Goor, M., Pot, B., Falsen, E., Jantzen, E., Hoste, B., Gillis, M., Kersters, K., Auling, G., and De Ley, J.: Hydrogenophaga, a new genus of hydrogen-oxidizing bacteria that includes Hydrogenophaga flava comb. nov. (formerly Pseudomonas flava), Hydrogenophaga palleronii (formerly Pseudomonas palleronii), Hydrogenophaga pseudoflava (formerly Pseudomonas pseudoflava and “Pseudomonas carboxydoflava”), and Hydrogenophaga taeniospiralis (formerly Pseudomonas taeniospiralis), Int. J. Syst. Evol. Microbiol., 39, 319–333, https://doi.org/10.1099/00207713-39-3-319, 1989.
Wu, X. Y., Zheng, G., Zhang, W. W., Xu, X. W., Wu, M., and Zhu, X. F.: Amphibacillus jilinensis sp. nov., a facultatively anaerobic, alkaliphilic bacillus from a soda lake, Int. J. Syst. Evol. Microbiol., 60, 2540–2543, https://doi.org/10.1099/ijs.0.018259-0, 2010.
Yanagawa, K., Nunoura, T., McAllister, S., Hirai, M., Breuker, A., Brandt, L., House, C., Moyer, C. L., Birrien, J. L., Aoike, K., and Sunamura, M.: The first microbiological contamination assessment by deep-sea drilling and coring by the D/V Chikyu at the Iheya North hydrothermal field in the Mid-Okinawa Trough (IODP Expedition 331), Front. Microbiol., 4, 327, https://doi.org/10.3389/fmicb.2013.00327, 2013.
Yang, C. X., Liu, Y. P., Bao, Q. H., Feng, F. Y., Liu, H. R., Zhang, X. J., and Zhao, Y. L.: Mongoliitalea lutea gen. nov., sp. nov., an alkaliphilic, halotolerant bacterium isolated from a haloalkaline lake, Int. J. Syst. Evol. Microbiol., 62, 647–653, https://doi.org/10.1099/ijs.0.031286-0, 2012.
Zavarzina, D. G., Sokolova, T. G., Tourova, T. P., Chernyh, N. A., Kostrikina, N. A., and Bonch-Osmolovskaya, E. A.: Thermincola ferriacetica sp. nov., a new anaerobic, thermophilic, facultatively chemolithoautotrophic bacterium capable of dissimilatory Fe (III) reduction. Extremophiles, 11, 1–7, https://doi.org/10.1007/s00792-006-0004-7, 2007.
Zhang, G., Dong, H., Xu, Z., Zhao, D., and Zhang, C.: Microbial diversity in ultra-high-pressure rocks and fluids from the Chinese Continental Scientific Drilling Project in China, Appl. Environ. Microbiol., 71, 3213–3227, https://doi.org/10.1128/AEM.71.6.3213-3227.2005, 2005.
Zhang, G., Dong, H., Jiang, H., Xu, Z., and Eberl, D. D.: Unique microbial community in drilling fluids from Chinese continental scientific drilling, Geomicrobiol. J., 23, 499–514, https://doi.org/10.1080/01490450600875860, 2006.